A radiomics nomogram for preoperatively predicting prognosis of patients in hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common cancer and ranks second as a major cause of cancer-related deaths globally (1,2). Moreover, its incidence has continuously increased in recent years. Approximately 850,200 new cases of HCC are diagnosed annually worldwide, and more than half of these patients are in China (3). Almost 70–90% of HCC cases are related to hepatitis B virus (HBV) infection, which is highly endemic in Asia-Pacific regions, particularly China (4). Partial hepatectomy is among the potential curative therapies in limited patients. However, the 5-year recurrence rate of HCC complicates 70% of cases after surgical therapies, and the overall time is still unsatisfactory (5,6). As such, screening patients who would benefit more from liver resection surgery is imperative.
Although the Barcelona Clinic Liver Cancer (BCLC) stage for HCC plays a crucial role in estimating prognosis, by facilitating treatment stratification, it may not be sufficiently precise (7,8). Moreover, studies have reported that various clinical risk factors, such as aspartate transaminase (AST), alpha-fetoprotein (AFP), and microvascular invasion were associated with poor survival (9-12). However, the clinical utility of these factors is limited. Thus, to facilitate individualized treatment strategies, a new tool to accurately identify patients who would have poor prognosis after partial hepatectomy is urgently needed.
Radiomics is an emerging and promising field that involves the extraction of several quantitative features from digital images, such as magnetic resonance imaging (MRI) or computed tomography (CT) (13,14). Several studies have determined the relationship between radiomics feature and the underlying pathophysiology (15-17). By converting medical images into high-dimensional and extractable data, radiomics algorithms provide an unprecedented opportunity to improve decision-making in oncology at a low-cost and in a noninvasive pattern, which helps oncologists to deliver more individualized medical care that considers phenotypic patient subtypes. A recent study reported that a radiomics nomogram can predict preoperative lymph node metastasis in patients with colorectal cancer (18). Previous studies have also indicated that biomarkers based on quantitative radiomics features are related to clinical prognosis and underlying genomic patterns across a range of cancer types (19,20). Besides, a recent study demonstrated that a radiomics signature, based on CT texture assessments, was a predictive biomarker for 2-year recurrence in patients with HCC (21). However, the association between radiomics signature and disease-free survival (DFS) in HBV-related HCC patients after partial hepatectomy has not been reported yet (22,23).
The present study aimed to develop and validate a radiomics model of personalized, non-invasive evaluation of DFS in HCC patients after hepatectomy treatment.
Methods
Patients
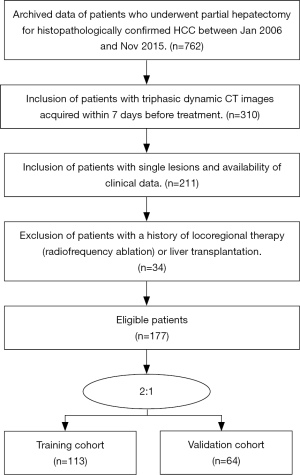
This retrospective study was approved by our institutional review board and Ethical Committee (NFEC-201208-K3). Informed consents were signed from patients or family members. Our study included consecutive patients in our center between Jan 2006 and Nov 2013 who met the following criteria: (I) pathologically proven HCC; (II) receive partial hepatectomy; (III) availability of 3-phase dynamic CT imaging within 7 days before surgery; and (IV) complete clinical and follow-up data. Besides, the exclusion criteria were as follows: (I) locoregional therapies, such as radiofrequency ablation and radiotherapy, or liver transplantation; (II) other liver malignant tumor; (III) two or more tumor diseases. We randomly divided the included patients into a training cohort and a validation cohort in a 2:1 ratio. Pretreatment clinical characteristics were collected from electronic medical records. Institutional review board approval was achieved for this study. Recruitment pathways for patients and flowchart for the process of model development and validation were shown in Figures S1,S2, respectively.
Follow-up
DFS and overall survival (OS) were the primary and secondary end points of this study, respectively. DFS was defined as the time from the day of the partial hepatectomy until either the date of relapse, death, or the patient was last known to be free of recurrence (censored). Meanwhile, OS was defined as the time from surgery treatment to death from any cause. Follow-up visits were conducted every 3 months during the first 2 years, and every 6 months for the next 3–5 years. All local recurrences were confirmed via MRI or CT at our center.
CT acquisition, region-of-interest segmentation, and radiomic feature extraction
Preoperative CT images were collected via a Picture Archiving and Communication System (PACS; Nanfang Hospital Network Center, China). More detailed information is shown in the Supplementary information.
Tumor regions of interest (ROIs) were semi-automatically segmented in the largest cross-sectional area using the IBEX software package (open source, source-code version) (24). Texture extraction was performed using IBEX software In total, 980 imaging features were extracted from the hepatic-arterial and portal-venous images, including intensity direct, intensity histogram, gray level run length matrix (GLRLM), gray level co-occurrence matrix (GLCM), neighbor intensity difference, and shape features. More information about the methodology for radiomics feature extraction can be found in Supplementary Method. Tumor ROIs were independently and manually segmented by an abdominal radiologist who had 12 years’ experience (reader 1) and another senior radiologist who had 22-year experience (reader 2). The both radiologists were blinded to the pathological results of the patients.
Inter- and intra-observer reproducibility analysis
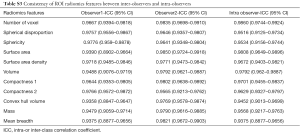
Inter-observer and intra-observer reproducibility of ROI segmentation and radiomic feature extraction were assessed using intra-and inter-class correlation coefficients (ICCs) in portal-venous images of 44 consecutive patients. To evaluate the intra-observer reproducibility, 2 radiologists independently segmented the ROIs of the 44 cases. Both readers generated the radiomic features twice with at least one-week interval between two readings to assess the inter-observer reproducibility. An ICC greater than 0.75 indicates a good agreement of the feature extraction. These values of many radiomic features, which describe the shape and size of the ROIs, can also be applied for evaluating the overall inter- and intra-observer agreement of the ROIs.
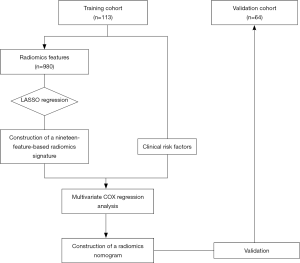
Feature selection and radiomics signature development
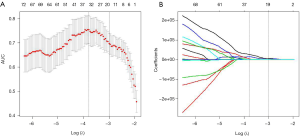
Using the least absolute shrinkage and selection operator (LASSO) algorithm, we built a logistic model and selected the λ in the smallest cross-validation error. Finally, a Rad-score formula was defined based on the 19 selected features. A radiomics signature was then constructed via the Rad-score. Using Kaplan-Meier survival analysis, we evaluated the potential relation between the radiomics signature and prognosis (DFS and OS) in the training and validation cohorts. The optimal cut-off value of Rad-score was determined using X-tile in DFS analysis.
Development and validation of the radiomics nomogram
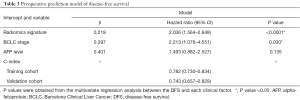
Multivariate regression analysis was performed to find a radiomics nomogram as a quantitative model to predict DFS. Candidate predictors of DFS were radiomics signature, BCLC stage, and AFP level.
The performance of the nomogram was evaluated in the internal validation cohort. DFS was then evaluated considering the total points as a factor in the Cox regression analysis. Finally, the C-index and calibration curves were derived using Cox regression analysis. Harrell’s C-index was evaluated to quantify the discrimination capability of the radiomics nomogram in the training cohort. The radiomics nomogram was internally validated using 10,000 bootstrap samples to achieve an optimism corrected performance.
Clinical utility
To evaluate the clinical utility of the nomogram, decision curve analysis (DCA) was used to quantify the probabilities of net benefits at different threshold in all patients.
Statistical analysis
All statistical analyses were performed via R statistical software version 3.3.3 (R Core Team, 2017). The “glmnet” package was used to perform the LASSO algorithm. The nomogram and the calibration curve plot were created using the “rms” package. The result of the Kaplan-Meier survival analysis was plotted using the “survminer” package, while that of the DCA was plotted using the “dca.R” package. A two-sided P value of <0.05 was considered statistically significant.
Results
Clinical characteristics
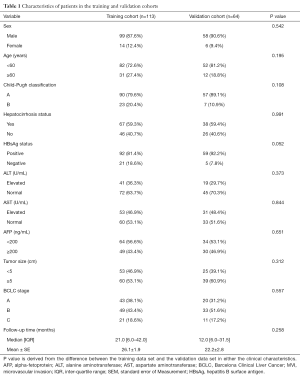
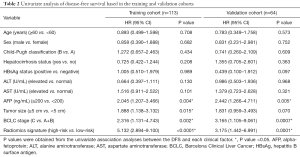
One hundred and seventy-seven patients were finally included in our study, among which 113 patients were allocated to the training cohort and 61 to the validation cohort. The study flowchart for patients in our study is shown in Figure S2, with the baseline clinical characteristics of the training and validation cohorts summarized in Table 1. In the final follow-up, 99 patients (55.9%) had confirmed disease recurrence or died. The mean and median DFS were 22.1 and 16.0 months, respectively. Clinical and follow-up data were not significantly different between the training and validation cohorts (P=0.052–0.991) (Table 1).
Full table
Radiomics feature extraction and radiomics signature construction
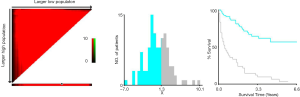
Flowchart for the radiomics feature extraction process was shown. A total of 980 features were extracted from the hepatic arterial and portal venous CT images and were finally reduced to 19 features (13 from hepatic-arterial images and 6 from portal-venous images) with the LASSO regression model (Figure 1). Based on the radiomic features describing the shape and size of the ROIs, the intra- and the inter-observer ICCs ranged from 0.937 to 0.986 and from 0.935 to 0.985, respectively, indicating favorable intra- and inter-observer feature extraction reproducibility (Table S3).
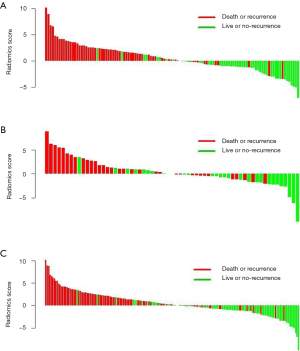
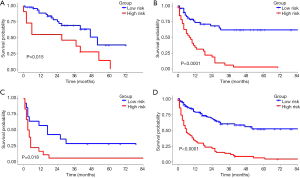
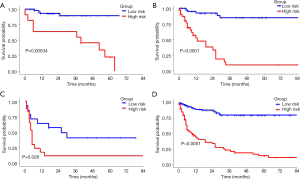
As a result, the radiomics signature, including 19 features was significantly associated with relapse or death in the training cohort (Figure 2). The optimal cut-off value of the Rad-score was 1.32 using the X-tile software in the DFS analysis Figure S3. In the training and validation cohorts, patients with a high Rad-score showed poorer DFS than those with a low Rad-score (HR, 5.132; 95% CI: 2.894–9.100; P<0.0001 and HR, 3.175; 95% CI: 1.442–6.991; P=0.00013, respectively). Furthermore, patients with a high Rad-score also showed poorer OS than those with a low Rad-score (HR, 7.329; 95% CI: 7.352–14.320; P<0.0001 and HR, 6.849; 95% CI: 2.350–19.970; P<0.0001 for both cohorts). The distributions of the Rad-scores and DFS status in the two cohorts and all patients are shown in Figure S4. Meanwhile, the results of the sub-analysis of the survival in BCLC-A, BCLC-B, and BCLC-C stages were shown in Figures S5,S6.

Development and validation of the radiomics model
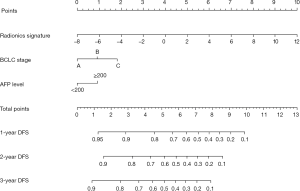
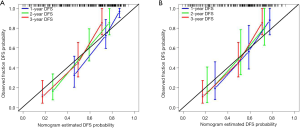
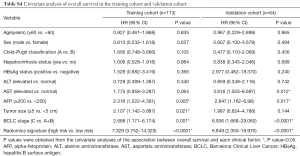
Univariate analysis indicated that the radiomics signature, AFP level, and BCLC stage were significantly associated with DFS or OS both in the training and validation cohorts (Table 2,S4). Additionally, the multivariate Cox regression analysis showed that the radiomics signature and BCLC stage were independent predictors of prognosis (Table 3). Notably, AFP level was also an important predictor. Thus, a radiomics nomogram was developed based on the radiomics signature, AFP level, and BCLC stage (Figure 3). This model showed favorable C-indexes of 0.782 (95% CI: 0.730–0.834) and 0.743 (95% CI: 0.657–0.829) for the training and the validation cohorts, respectively (Table 3). Besides, a good agreement between the nomogram-estimated probability and actual DFS status was noted and shown in the calibration curves (Figure 4A,B).
Full table
Full table
Clinical utility
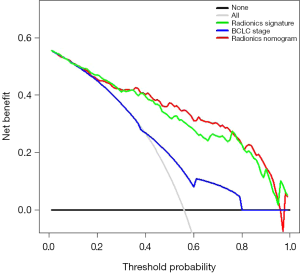
The DCA was estimated on the radiomics signature, BCLC stage, and radiomics nomogram, as shown in Figure 5. The decision curve of the radiomics signature and radiomics nomogram showed relatively good performance for the model regarding clinical application. It was indicated from the DCA curve that when the threshold probability of a patient or doctor is 15%, more benefit would be acquired using radiomics nomogram or radiomics signature than either treat-all or treat-none strategies. The probability of achieving DFS ranged from 38% to 80%. Both radiomics nomogram and radiomics signature had higher overall net benefits than the single BCLC stage. Thus, either radiomics nomogram or radiomics signature is more accurate for discriminating patients who could benefit from partial hepatectomy than traditional BCLC staging system.
Discussion
In the present study, 82.31% of patients had HBV infection, which is the crucial pathogenic factor of HCC in Asia-Pacific regions (4). Based on pre-treatment CT images of those patients, we developed a radiomics nomogram and validated this model as a tool for noninvasive, individualized prediction of DFS. The easy-to-use radiomics nomogram had an excellent performance and could help health care professionals in clinical decision making.
The precise estimation of DFS via conventional CT images remains challenging in clinical settings. Currently, different staging systems combining image features (e.g., tumor size and number) and clinical risk factors may also perform well (25-28). However, their levels of accuracy are unsatisfactory. We used a radiomics analysis model to transform images into high-dimensional radiomics features, which were used to estimate patient prognosis. We extracted 490 features from each CT image and were then reduced to 19 radiomics features using the LASSO algorithm and chosen via 10-fold cross-validation to develop a radiomics signature. We found that patients with high Rad-score showed significantly poorer DFS or OS than those with low Rad-score. Previous studies also showed that the radiomics signature could be used to predict the prognosis of patients with advanced nasopharyngeal carcinoma or early non-small cell lung cancer (29-32). Similar to previous studies, our study indicated that this novel method predicting prognosis via radiomics features extracted from CT images may help clinicians accurately predict the prognosis of patients with HCC who underwent partial hepatectomy.
The BCLC staging system is a standard tool used for clinical management of HCC and can be used to determine the appropriate therapeutic strategy (33-36). However, treatment recommendations for patients with HCC vary in different staging systems. For example, the BCLC staging classification recommends only transarterial chemoembolization for patients categorized with stage B HCC, while the Japan Society of Hepatology stipulates that partial hepatectomy is feasible for such patients with acceptable prognoses. Therefore, the indications for partial hepatectomy differ among several staging systems. Subgroup analysis in our study showed that the Rad-score generated from the radiomics signature can easily identify stage B patients (classified according to the BCLC system) who would benefit more from the surgery. Interestingly, a small proportion of stage A patients (classified according to the BCLC system) with high Rad-score who underwent partial hepatectomy still have poor DFS, indicating that these patients should be given more attention on re-examination. Moreover, although most stage C patients showed poor prognosis after partial hepatectomy, several patients with low Rad-score also obtained benefit from surgery.
In addition, previous studies have reported that several clinical risk factors, such as high AFP levels and large tumor size, were associated with poor prognosis in HCC patients (37,38). Univariate analysis of DFS and OS in the training and validation cohorts indicated that most clinical risk factors (e.g., age, sex, pre-treatment Child-Pugh classification, pre-treatment hepatocirrhosis status, pre-treatment HBsAg status, pre-treatment alanine aminotransferase, and pre-treatment AST) were not significantly associated with prognosis. However, similar to previous studies, high AFP levels were also significantly associated with poor DFS and OS in both the training and validation cohorts (39-41). Tumor size was associated with DFS and OS in the training cohort, while it was not associated with prognosis in the validation set.
Considering the above factors, we developed a radiomics nomogram that incorporated the radiomics signature, BCLC stage, and AFP level. The nomogram can be a tool for developing an individualized treatment strategy. To the best of our knowledge, the use of pre-treatment radiomics signature for DFS prediction has never been reported. The radiomics model indicated favorable consistency in the training cohort, and the outcome was verified in the validation cohort (C-index, 0.782 and 0.743, respectively), which is shown as a calibration curve. Next, we plotted the decision curve based on the radiomics nomogram, radiomics signature, and BCLC stage in all the patients. DCA indicated that using the radiomics nomogram to estimate DFS when a threshold probability of a patient or doctor is 18%, higher benefits obtained than the overall or no treatment strategy in our study. The probability of achieving DFS with partial hepatectomy in patients classified according to BCLC stage ranges from 38–80%. We also found that both the radiomics nomogram and radiomics signature had higher overall net benefits than BCLC stage alone.
Our study had two limitations. First, the numbers of patients with HCC was relatively small. Second, the data were collected from a single institution. If multicenter cohorts are enrolled and the collective data of different parameters are used, our model may perform differently. Thus, much larger datasets must be collected from multiple centers, and the robustness and reproducibility of our proposed radiomics model needs to be investigated. In summary, our model that comprised of radiomics features, BCLC stage, and AFP levels can serve as a non-invasive and preoperative tool to predict DFS in patients with HBV-related HCC undergoing partial hepatectomy. This model can be used to decide more precise clinical treatments.
Parameters for computed tomography image acquisition
All patients underwent pre-treatment multi-detector row CT (MDCT) scans (SOMATOM and 256-iCT). The acquisition parameters were shown in Table S1.
CT acquisition
Contrast-enhanced computed tomography (CECT) was performed at our hospital with one of the following MDCT scanners: SOMATOM (Siemens Medical Systems, Erlangen, Germany), or Brilliance iCT 256 (Philips Healthcare, Cleveland, OH, USA). The scanning parameters are shown in Table S1. After routine CT scanning, a contrast agent (Ultravist 370, Bayer Schering Pharma, Berlin, Germany, 1.0 mL/kg) was injected into the antecubital vein at the rate of 2.0–3.0 mL/s via an injector (Ulrich CT Plus 150, Ulrich Medical, Ulm, Germany); a CECT was performed immediately after injection. The hepatic-arterial and portal-venous phase CT images were acquired at 30 and 60 s, respectively. Preoperative CT images were collected on the Picture Archiving and Communication System (PACS; Nanfang Hospital Network Center, China), with an optimal window setting adjustment (window width: 300, window level: 50).
Supplementary methods
In this study, a total number of 980 candidate radiomics features were derived from the arterial and portal-venous phase images with 490 for each. IBEX software package on MATLAB 2014b (Math Works, Natick, MA, USA) was used for feature extraction. The 980 features included intensity direct, intensity histogram, gray level run length matrix (GLRLM), gray level co-occurrence matrix (GLCM), neighbor intensity difference, and shape features.
Nineteen features that were most associated with DFS via a least absolute shrinkage and selection operator (LASSO) method. The features were shown in Table S2.
Radiomics score (Rad-score) calculation formula
Rad-score=−2.754e+04
−4.219e−01*A_NeighborIntensityDifference25_Texture Strength
−1.620e−02*A_IntensityHistogram_35PercentileArea
−5.090e−01 *A_IntensityHistogram_Kurtosis
+5.069e−02*A_IntensityDirect_LocalStdMax
−0*A_IntensityDirect_Kurtosis
−2.375e-02*A_IntensityDirect_GlobalMax
+1.653e+04*A_GLCM 25_90-1InverseDiffMomentNorm
+1.105e+04*A_GLCM 25_0-7InverseDiffMomentNorm
+6.111e+00*A_GLCM 25_135-7Correlation
−9.453e−01*A_GLCM 25_90-7Correlation
−6.162e+00*A_GLCM 25_-333-7Correlation
+3.916e+00*A_GLCM 25_45-4Correlation
+9.464e−01*A_GLCM 25_90-7ClusterShade
−2.757e−03*P_IntensityHistogram_10PercentileArea
+1.010e+00*P_IntensityDirect_LocalEntropyMin
−2.321e+01*P_GLCM 25_135-7InformationMeasureCorr1
+1.496e+01*P_GLCM 25_45-1Energy
−2.992e−01*P_GLCM 25_135-7DifferenceEntropy
−4.086e−03*P_GLCM 25_0-1ClusterProminence
−P and A indicate that the features were retrieved from portal and arterial phase CT images, respectively.

Full table

Full table
Full table
Full table
Acknowledgments
Funding: This work was supported by the National Nature Science Foundation of China (Grant Nos. 81372283, 81472711, 81401180, 81672756, and 91540111), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2015), and the Natural Science Foundation of Guangdong Province (Grant No. 2014A030311013).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.06.18). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This retrospective study was approved by our institutional review board and Ethical Committee (NFEC-201208-K3). Informed consents were signed from patients or family members.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Omata M, Cheng AL, Kokudo N, et al. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatol Int 2017;11:317-70. [Crossref] [PubMed]
- Ding J, Wang H. Multiple interactive factors in hepatocarcinogenesis. Cancer Lett 2014;346:17-23. [Crossref] [PubMed]
- Ding XX, Zhu QG, Zhang SM, et al. Precision medicine for hepatocellular carcinoma: driver mutations and targeted therapy. Oncotarget 2017;8:55715-30. [PubMed]
- Lei Z, Li J, Wu D, et al. Nomogram for Preoperative Estimation of Microvascular Invasion Risk in Hepatitis B Virus-Related Hepatocellular Carcinoma Within the Milan Criteria. JAMA Surg 2016;151:356-63. [Crossref] [PubMed]
- Grazi GL, Ercolani G, Pierangeli F, et al. Improved results of liver resection for hepatocellular carcinoma on cirrhosis give the procedure added value. Ann Surg 2001;234:71-8. [Crossref] [PubMed]
- Lim KC, Chow PK, Allen JC, et al. Microvascular invasion is a better predictor of tumor recurrence and overall survival following surgical resection for hepatocellular carcinoma compared to the Milan criteria. Ann Surg 2011;254:108-13. [Crossref] [PubMed]
- Bruix J, Sherman M, Llovet JM, et al. Clinical management of hepatocellular carcinoma. Conclusions of the Barcelona-2000 EASL conference. European Association for the Study of the Liver. J Hepatol 2001;35:421-30. [Crossref] [PubMed]
- European Association For The Study Of The L. European Organisation For R, Treatment Of C. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol 2012;56:908-43. [Crossref]
- Banerjee S, Wang DS, Kim HJ, et al. A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology 2015;62:792-800. [Crossref] [PubMed]
- Hirokawa F, Hayashi M, Asakuma M, et al. Risk factors and patterns of early recurrence after curative hepatectomy for hepatocellular carcinoma. Surg Oncol 2016;25:24-9. [Crossref] [PubMed]
- Hwang S, Song GW, Lee YJ, et al. Multiplication of Tumor Volume by Two Tumor Markers Is a Post-Resection Prognostic Predictor for Solitary Hepatocellular Carcinoma. J Gastrointest Surg 2016;20:1807-20. [Crossref] [PubMed]
- Sasaki K, Matsuda M, Ohkura Y, et al. Factors associated with early cancer-related death after curative hepatectomy for solitary small hepatocellular carcinoma without macroscopic vascular invasion. J Hepatobiliary Pancreat Sci 2014;21:142-7. [Crossref] [PubMed]
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Kuo MD, Gollub J, Sirlin CB, et al. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J Vasc Interv Radiol 2007;18:821-31. [Crossref] [PubMed]
- Kuo MD, Jamshidi N. Behind the numbers: Decoding molecular phenotypes with radiogenomics--guiding principles and technical considerations. Radiology 2014;270:320-5. [Crossref] [PubMed]
- Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- Li Y, Liu X, Xu K, et al. MRI features can predict EGFR expression in lower grade gliomas: A voxel-based radiomic analysis. Eur Radiol 2018;28:356-62. [Crossref] [PubMed]
- Huang YQ, Liang CH, He L, et al. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J Clin Oncol 2016;34:2157-64. [Crossref] [PubMed]
- Lao J, Chen Y, Li ZC, et al. A Deep Learning-Based Radiomics Model for Prediction of Survival in Glioblastoma Multiforme. Sci Rep 2017;7:10353. [Crossref] [PubMed]
- Zhang B, He X, Ouyang F, et al. Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett 2017;403:21-7. [Crossref] [PubMed]
- Zhou Y, He L, Huang Y, et al. CT-based radiomics signature: a potential biomarker for preoperative prediction of early recurrence in hepatocellular carcinoma. Abdom Radiol (NY) 2017;42:1695-704. [Crossref] [PubMed]
- Kiryu S, Akai H, Nojima M, et al. Impact of hepatocellular carcinoma heterogeneity on computed tomography as a prognostic indicator. Sci Rep 2017;7:12689. [Crossref] [PubMed]
- Chen S, Zhu Y, Liu Z, et al. Texture analysis of baseline multiphasic hepatic computed tomography images for the prognosis of single hepatocellular carcinoma after hepatectomy: A retrospective pilot study. Eur J Radiol 2017;90:198-204. [Crossref] [PubMed]
- Zhang L, Fried DV, Fave XJ, et al. IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys 2015;42:1341-53. [Crossref] [PubMed]
- Liang W, Shen G, Zhang Y, et al. Development and validation of a nomogram for predicting the survival of patients with non-metastatic nasopharyngeal carcinoma after curative treatment. Chin J Cancer 2016;35:98. [Crossref] [PubMed]
- Liang W, Zhang L, Jiang G, et al. Development and validation of a nomogram for predicting survival in patients with resected non-small-cell lung cancer. J Clin Oncol 2015;33:861-9. [Crossref] [PubMed]
- Wu J, Chen QX. Prognostic and predictive significance of tumor length in patients with esophageal squamous cell carcinoma undergoing radical resection. BMC Cancer 2016;16:394. [Crossref] [PubMed]
- Xiao HF, Zhang BH, Liao XZ, et al. Development and validation of two prognostic nomograms for predicting survival in patients with non-small cell and small cell lung cancer. Oncotarget 2017;8:64303-16. [Crossref] [PubMed]
- Huang Y, Liu Z, He L, et al. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology 2016;281:947-57. [Crossref] [PubMed]
- Li Q, Kim J, Balagurunathan Y, et al. CT imaging features associated with recurrence in non-small cell lung cancer patients after stereotactic body radiotherapy. Radiat Oncol 2017;12:158. [Crossref] [PubMed]
- Takeda K, Takanami K, Shirata Y, et al. Clinical utility of texture analysis of 18F-FDG PET/CT in patients with Stage I lung cancer treated with stereotactic body radiotherapy. J Radiat Res 2017;58:862-869. [Crossref] [PubMed]
- Zhang B, Tian J, Dong D, et al. Radiomics Features of Multiparametric MRI as Novel Prognostic Factors in Advanced Nasopharyngeal Carcinoma. Clin Cancer Res 2017;23:4259-69. [Crossref] [PubMed]
- El-Serag HB. Advances in the management of hepatocellular carcinoma. Clin Adv Hematol Oncol 2017;15:2-6. [PubMed]
- Mokdad AA, Hester CA, Singal AG, et al. Management of hepatocellular in the United States. Chin Clin Oncol 2017;6:21. [Crossref] [PubMed]
- Sastre J, Diaz-Beveridge R, Garcia-Foncillas J, et al. Clinical guideline SEOM: hepatocellular carcinoma. Clin Transl Oncol 2015;17:988-95. [Crossref] [PubMed]
- Yu SJ. A concise review of updated guidelines regarding the management of hepatocellular carcinoma around the world: 2010-2016. Clin Mol Hepatol 2016;22:7-17. [Crossref] [PubMed]
- Fu YP, Yi Y, Huang JL, et al. Prognostic Nomograms Stratify Survival of Patients with Hepatocellular Carcinoma Without Portal Vein Tumor Thrombosis After Curative Resection. Oncologist 2017;22:561-9. [Crossref] [PubMed]
- Kao WY, Su CW, Chiou YY, et al. Hepatocellular Carcinoma: Nomograms Based on the Albumin-Bilirubin Grade to Assess the Outcomes of Radiofrequency Ablation. Radiology 2017;285:670-80. [Crossref] [PubMed]
- Bruix J, Cheng AL, Meinhardt G, et al. Prognostic factors and predictors of sorafenib benefit in patients with hepatocellular carcinoma: Analysis of two phase III studies. J Hepatol 2017;67:999-1008. [Crossref] [PubMed]
- Shen JY, Li C, Wen TF, et al. Transplantation versus hepatectomy for HCC beyond the Milan criteria: A propensity score analysis. Int J Surg 2017;44:33-42. [Crossref] [PubMed]
- Silva JP, Gorman RA, Berger NG, et al. The prognostic utility of baseline alpha-fetoprotein for hepatocellular carcinoma patients. J Surg Oncol 2017;116:831-40. [Crossref] [PubMed]