
Circular RNA and exosomes in pancreatic cancer progression
Circular RNA (circRNA) is a type of noncoding RNA that forms a covalently closed continuous loop. CircRNAs were discovered in the 1980s but were thought to be an artifact of aberrant RNA splicing or specific to a few pathogens such as hepatitis delta virus (1). The development of next-generation sequencing and bioinformatics tools has led to the discovery of numerous endogenous circRNAs, the expression of which is considerably higher than previously thought. The expression levels of circRNAs differ among cell types and are often independent of those of the parent gene. Thus, circRNAs are not merely artifacts of aberrant RNA splicing but have unique functions. CiRS-7, one of the best studied circRNAs, has more than 70 seed sequences capable of binding to microRNA-7 and inhibiting its binding to the target mRNA. CiRS-7 functions as a competing endogenous RNA for microRNA-7 or as a microRNA-7 sponge (2). CircRNAs can also interact with RNA to control transcription (3) or translation (4,5). Furthermore, circRNAs have been implicated in the pathogenesis of various cancers, arteriosclerosis, and central nervous system diseases (6).
Exosomes are secreted vesicles 30 to 100 nm in diameter that are surrounded by a lipid bilayer membrane. Exosomes contain host-cell-derived RNA and protein and can be transported long distances to induce functional and physiological changes in target cells. Exosomes secreted by cancer cells play an important role in cancer progression and metastasis by inhibiting the activation of T cells and dendritic cells and inducing cancer cells to undergo epithelial-mesenchymal transition (7,8).
Exosomes contain microRNAs, tRNAs, mRNAs, and according to recent reports, circRNAs (9). In exosomes, circRNAs are enriched compared with the linear transcripts derived from their parent gene (9). Moreover, circRNAs are more abundant in exosomes than in the cell from which the exosomes originated. Colon, lung, stomach, breast, and cervical cancer cells secrete circRNA-containing exosomes. In exosomes present in the serum of patients with colorectal cancer, certain circRNAs were enriched compared with those in exosomes from healthy controls (9). Therefore, exosomal circRNAs are implicated in the pathogenesis and/or progression of cancer.
Li et al. first reported a role for exosomal circPED8A in pancreatic cancer. They performed microarray analysis and identified circ-PDE8A in exosomes secreted by Hs766T-L2 pancreatic cancer cells, which were established from liver metastatic tissues. The expression level of circ-PDE8A in pancreatic cancer tissue was related to lymphatic invasion, TNM stage, and a poor survival rate. In addition, the expression level of circPDE8A in exosomes from the plasma of pancreatic cancer patients was related to the progression and prognosis of pancreatic cancer. Moreover, circPDE8A acted as a competing endogenous RNA for microRNA-338 and induced invasive growth via the MACC, MET, ERK, and AKT pathways (Figure 1). Li and coworkers also established stably Hs766T-L2 cells stably overexpressing circPDE8A, and the exosomes secreted from these cells were rich in circPDE8A transcripts; co-culture of these exosomes with BxPC-3 pancreatic cancer cells resulted in upregulation of MACC expression in the BxPC-3 cells.
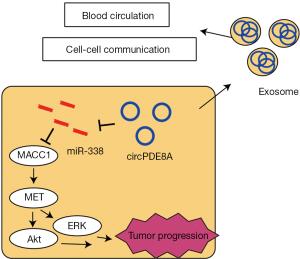
These novel findings imply that exosomal circPDE8A promotes tumor growth via the MACC, MET, ERK, and AKT pathways in a cell-autonomous manner and promotes the progression of pancreatic cancer in a paracrine manner. However, several questions remain to be resolved.
First, how are circRNAs enriched during exosome formation? Exosomes originate from various sizes of endoplasmic reticulum known as intraluminal vesicles, which are formed by budding from multivesicular bodies. One possibility is that circRNAs are present in the cytoplasm and passively included in exosomes during their formation. As circRNAs are more recalcitrant to exonuclease degradation than are linear RNAs, circRNAs may become enriched upon degradation of linear RNA. Alternatively, circRNAs may be actively transported from the cytoplasm into exosomes. CiRS-7 functions as an microRNA-7 sponge. Introduction of an microRNA-7 mimic into cells reduced the abundance of ciRS-7 in exosomes but increased that in the exosome-derived cells (9). In addition, the abundance of circRTN4 was increased in exosomes derived from DLD-1 colorectal cancer cells, which possess a KRAS mutation, compared with DKs-8 cells (wild type KRAS), whereas the intracellular abundance of circRTN4 was decreased in DLD-1 cells (10). Therefore, exosomal circRNA is independent of intracellular circRNA and may be actively transported between exosomes and the cytoplasm. However, the transporter involved has yet to be identified, and thus further research is needed.
Several circRNAs are associated with the growth of pancreatic cancer. For example, circRNA-100782 controls tumor growth via the IL-6/STAT3 pathway by acting as an microRNA-124 sponge (11). Silencing of the circRNA hsa_circ_0000977 suppresses the progression of pancreatic ductal adenocarcinoma by stimulating microRNA-874-3p and inhibiting PLK1 expression (12). Although circRNAs can promote cancer progression, their only known function is acting as microRNA sponges. Despite the discovery of numerous circRNAs, the functions of only a few have been identified. Thus, further studies are required to gain a complete understanding of the functions of circRNAs.
In this study, exosomal circPDE8A was shown to interact with pancreatic cancer cells. Exosomes interact with not only cancer cells but also fibroblasts, endothelial cells, and immune cells to promote tumor growth and invasion (7). Tumor-derived exosomes induce differentiation of cancer-associated fibroblasts, which play a major role in the establishment of the tumor microenvironment. Fibroblast-derived exosomes also support tumor growth, and exosome-mediated communication between tumor cells and fibroblasts is associated with cancer progression. Exosomal communication between tumor cells and endothelial cells regulates angiogenesis and promotes tumor growth, while that between tumor cells and immune cells modulates the host immune response and promotes inflammation. Thus, exosomes play an important role in tumor angiogenesis, inflammation, and immune reconstitution. Further research is needed to investigate the effect of exosomal circRNAs on the tumor microenvironment.
In this study, a high level of exosomal circPDE8A in plasma, as determined by quantitative reverse-transcription polymerase chain reaction (qRT-PCR), was correlated with the progression and prognosis of pancreatic cancer. The housekeeping genes β-actin and GAPDH are typically used as an internal control in qRT-PCR, but whether they are appropriate for quantification of exosomal circRNAs in plasma is unclear.
Several issues regarding the relationship between exosomal circRNAs and pancreatic cancer remain to be resolved. The findings of this study indicate that circRNAs and exosomes are involved in the progression of pancreatic cancer, implying their potential as biomarkers and therapeutic targets. Therefore, further research on exosomal circRNAs will enhance our understanding of the pathology of pancreatic cancer.
Acknowledgments
Funding: This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan (#16H05149, #16KT0109, and #15H04807 to M Otsuka, M Otsuka, and K Koike), supported by the Research Program on Hepatitis from Japan Agency for Medical Research and Development, AMED (to M Otsuka, #JP18fk0210214), and by the Project for Cancer Research And Therapeutic Evolution (P-CREATE) from AMED (to M Otsuka, #JP19cm0106602).
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Chunlin Ou (Cancer Research Institute of Central South University, Changsha, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.08.03). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. [Crossref] [PubMed]
- Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 2017;66:22-37.e9. [Crossref] [PubMed]
- Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell 2017;66:9-21.e7. [Crossref] [PubMed]
- Lei K, Bai H, Wei Z, et al. The mechanism and function of circular RNAs in human diseases. Exp Cell Res 2018;368:147-58. [Crossref] [PubMed]
- Maia J, Caja S, Strano Moraes MC, et al. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front Cell Dev Biol 2018;6:18. [Crossref] [PubMed]
- Syn N, Wang L, Sethi G, et al. Exosome-Mediated Metastasis: From Epithelial-Mesenchymal Transition to Escape from Immunosurveillance. Trends Pharmacol Sci 2016;37:606-17. [Crossref] [PubMed]
- Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015;25:981-4. [Crossref] [PubMed]
- Dou Y, Cha DJ, Franklin JL, et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep 2016;6:37982. [Crossref] [PubMed]
- Chen G, Shi Y, Zhang Y, et al. CircRNA_100782 regulates pancreatic carcinoma proliferation through the IL6-STAT3 pathway. Onco Targets Ther 2017;10:5783-94. [Crossref] [PubMed]
- Huang WJ, Wang Y, Liu S, et al. Silencing circular RNA hsa_circ_0000977 suppresses pancreatic ductal adenocarcinoma progression by stimulating miR-874-3p and inhibiting PLK1 expression. Cancer Lett 2018;422:70-80. [Crossref] [PubMed]


