
The urea cycle enzymes act as metabolic suppressors in clear cell renal cell carcinoma
Renal cell carcinoma (RCC) comprises about 3–4% of adult malignancies and according to recent estimates, over 65,000 new cases will be diagnosed in 2018 (3.8% of all new cancer cases), and nearly 15,000 patients will die of this tumor (2.5% of all cancer deaths) in the United States (1).
Although patients with low stage, localized RCC have an excellent prognosis after surgical treatment (92.6% of these patients are still alive 5 years after the diagnosis), nearly 30% are diagnosed with metastatic disease, and their 5-year survival is less than 12% (1-3).
Recent studies have investigated novel molecular mechanisms involved in the RCC pathogenesis, and have identified potential biomarkers with a role in early diagnosis, risk assessment, and outcome prediction. Different molecular markers such as CA 15-3, αKlotho, RKIP and many metabolic enzymes have been investigated, but none of these factors is used in the clinical management of kidney cancer patients (4-11).
The recent introduction of high-throughput screening has led to an in-depth molecular characterization of different human cancers including urological tumors, together with the identification of novel pathogenic mechanisms and potential therapeutic targets.
This multi-omics approach has confirmed and extended the Otto Warburg hypothesis that cancer cells hijack and remodel existing metabolic pathways to promote cell survival and proliferation.
Many clinical studies have shown that RCC is fundamentally a metabolic disease (12). In fact, some clinical conditions characterized by an altered metabolism, such as obesity, diabetes and chronic kidney disease, are common risk factors for RCC (13-15). Moreover, molecular analyses have revealed that in the RCC tumor cell metabolism a program of metabolic remodeling is activated, characterized by a Warburg effect-like state, a rerouting of the sugar metabolism toward the pentose phosphate pathway, a reduced tricarboxylic acid (TCA) cycle activity, increased glutaminolysis and fatty acid accumulation (16-18).
In recent years, a series of metabolic adaptations involving the accumulation of uncommon oncometabolites has been described (9,19-21). In this scenario, a recent study showed that in clear cell RCC (ccRCC)—the most common kidney cancer subtype—the expression of multiple urea cycle enzymes was strongly repressed, suggesting a tumor suppressant role in normal physiological conditions (22).
Urea cycle activity occurs in the liver and kidney, to prevent the accumulation of toxic ammonia in the organism (Figure 1). The first compound to enter the cycle is carbamoyl phosphate, generated from ammonia in the mitochondrion by carbamoyl ph osphate synthetase (CPS). The cycle is characterized by four enzymatic steps. Firstly, carbamoyl phosphate donates its carbamoyl group to ornithine to form citrulline (step 1). Ornithine has a similar role to that of oxaloacetate in the TCA cycle, accepting material at each turn of the cycle. The reaction is catalyzed by ornithine transcarbamoylase (OTC), and citrulline shifts from the mitochondrion to the cytosol. The second amino group now enters from aspartate, by a condensation reaction between aspartate and citrulline, forming argininosuccinate (step 2). This cytosolic reaction is catalyzed by arginine-succinate synthetase 1 (ASS1). The arginine-succinate is then cleaved by arginine-succinate lyase (ASL) (step 3) to form arginine and fumarate, and the latter enters mitochondria to join the pool of TCA cycle intermediates. In the last reaction of the urea cycle (step 4), the cytosolic enzyme arginase 2 (ARG2) cleaves arginine to generate urea and ornithine. Ornithine is then transported into the mitochondrion to start another round of the urea cycle.
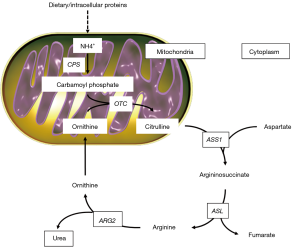
In this setting, Ochocki and colleagues (22) demonstrated that ccRCC are characterized by alterations in ammonia metabolism, in association with downregulation of multiple urea cycle enzymes including ARG2, ASS1 and ASL.
In particular, these authors showed that ARG2 and ASS1, when re-expressed in ccRCC cancer cells, suppressed tumor growth in vitro and in vivo. Moreover, to better define the mechanisms involved in tumor suppressor activity of urea cycle enzymes, the metabolomics profile related to ARG2 enzyme activity, that catalyzes the last reaction in the cycle with the production of urea and ornithine, was analyzed.
Ornithine can enter in multiple biochemical pathways apart from the urea cycle:
- Conversion to glutamate-γ-semialdehyde by mitochondrial ornithine aminotransferase (OAT) for the production of both glutamine and proline;
- Decarboxylation by ornithine decarboxylase (ODC) to synthetize polyamine.
Both these reactions require pyridoxal-5'-phosphate (PLP), a vitamin B6 derivative cofactor involved in a variety of metabolic reactions that are critical for glycogen production and amino acids synthesis. The authors demonstrated that ARG2 inhibits ccRCC tumor cell proliferation, through depletion of the PLP pool, hence reducing amino acids synthesis. The second mechanism involves the excessive production of polyamines putrescine and spermidine. In particular, using metabolic tracing experiments, it was shown that due to ARG2 activity, polyamines accumulate and cause cellular toxicity. Taken together, these findings suggest that in ccRCC a program of repression of urea cycle enzymes exists, with the aim of sustaining biomass expansion by two main mechanisms: maintaining elevated levels of PLP for amino acids homeostasis, and avoiding toxic polyamines concentrations.
A reduced expression of urea cycle enzymes has been reported also in other tumors such as sarcomas (23-25). Interestingly, Rabinovich and colleagues (25) showed, for the first time, a metabolic connection between the urea cycle enzymes and pyrimidine nucleotides, that are essential compounds for DNA synthesis. In particular, in different cancer cell lines an ASS1 deficiency was demonstrated to support tumor proliferation by activating carbamoyl-phosphate synthetase 2, aspartate transcarbamoylase, and dihydroorotase (CAD), increasing aspartate levels for pyrimidine synthesis.
These studies confirm the fundamental role of metabolic reprogramming in cancer cells, and lay the basis for the identification of novel therapeutic targets in ccRCC. Ochocki et al. have shown, for the first time, the central role of the ammonia metabolism in ccRCC and their findings offer fundamental support of the concept that study of the cancer metabolome may give rise to innovative approaches to patient risk stratification and drug development.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Dr. Peng Zhang (Department of Urology, Zhongnan Hospital of Wuhan University, Wuhan, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.08.07). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Battaglia M, Lucarelli G. The role of renal surgery in the era of targeted therapy: the urologist′s perspective. Urologia 2015;82:137-8. [Crossref] [PubMed]
- Di Lorenzo G, De Placido S, Pagliuca M, et al. The evolving role of monoclonal antibodies in the treatment of patients with advanced renal cell carcinoma: a systematic review. Expert Opin Biol Ther 2016;16:1387-401. [Crossref] [PubMed]
- Lucarelli G, Ditonno P, Bettocchi C, et al. Diagnostic and prognostic role of preoperative circulating CA 15-3, CA 125, and beta-2 microglobulin in renal cell carcinoma. Dis Markers 2014;2014:689795 [Crossref] [PubMed]
- Gigante M, Lucarelli G, Divella C, et al. Soluble Serum αKlotho Is a Potential Predictive Marker of Disease Progression in Clear Cell Renal Cell Carcinoma. Medicine (Baltimore) 2015;94:e1917 [Crossref] [PubMed]
- Papale M, Vocino G, Lucarelli G, et al. Urinary RKIP/p-RKIP is a potential diagnostic and prognostic marker of clear cell renal cell carcinoma. Oncotarget 2017;8:40412-24. [Crossref] [PubMed]
- Gigante M, Pontrelli P, Herr W, et al. miR-29b and miR-198 overexpression in CD8+T cells of renal cell carcinoma patients down-modulates JAK3 and MCL-1 leading to immune dysfunction. J Transl Med 2016;14:84. [Crossref] [PubMed]
- Lucarelli G, Rutigliano M, Sanguedolce F, et al. Increased expression of the autocrine motility factor is associated with poor prognosis in patients with clear cell-renal cell carcinoma. Medicine (Baltimore) 2015;94:e2117 [Crossref] [PubMed]
- Lucarelli G, Rutigliano M, Ferro M, et al. Activation of the kynurenine pathway predicts poor outcome in patients with clear cell renal cell carcinoma. Urol Oncol 2017;35:461.e15-461.e27. [Crossref] [PubMed]
- Farber NJ, Kim CJ, Modi PK, et al. Renal cell carcinoma: the search for a reliable biomarker. Transl Cancer Res 2017;6:620-32. [Crossref] [PubMed]
- Modi PK, Farber N, Singer E. Precision oncology: identifying predictive biomarkers for the treatment of metastatic renal cell carcinoma. Transl Cancer Res 2016;5:S76-80. [Crossref] [PubMed]
- Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol 2010;7:277-85. [Crossref] [PubMed]
- Tahbaz R, Schmid M, Merseburger AS. Prevention of kidney cancer incidence and recurrence: lifestyle, medication and nutrition. Curr Opin Urol 2018;28:62-79. [PubMed]
- Vavallo A, Simone S, Lucarelli G, et al. Pre-existing type 2 diabetes mellitus is an independent risk factor for mortality and progression in patients with renal cell carcinoma. Medicine (Baltimore) 2014;93:e183 [Crossref] [PubMed]
- Breda A, Lucarelli G, Rodriguez-Faba O, et al. Clinical and pathological outcomes of renal cell carcinoma (RCC) in native kidneys of patients with end-stage renal disease: a long-term comparative retrospective study with RCC diagnosed in the general population. World J Urol 2015;33:1-7. Erratum in: World J Urol 2015;33:9 Luccarelli, Giuseppe [corrected to Lucarelli, Giuseppe]. [Crossref] [PubMed]
- Hakimi AA, Reznik E, Lee CH, et al. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell 2016;29:104-16. [Crossref] [PubMed]
- Lucarelli G, Galleggiante V, Rutigliano M, et al. Metabolomic profile of glycolysis and the pentose phosphate pathway identifies the central role of glucose-6-phosphate dehydrogenase in clear cell-renal cell carcinoma. Oncotarget 2015;6:13371-86. [Crossref] [PubMed]
- Lucarelli G, Ferro M, Battaglia M. Multi-omics approach reveals the secrets of metabolism of clear cell-renal cell carcinoma. Transl Androl Urol 2016;5:801-3. [Crossref] [PubMed]
- von Rundstedt FC, Rajapakshe K, Ma J, et al. Integrative pathway analysis of metabolic signature in bladder cancer: a linkage to the cancer genome atlas project and prediction of survival. J Urol 2016;195:1911-9. [Crossref] [PubMed]
- Lucarelli G, Rutigliano M, Galleggiante V, et al. Metabolomic profiling for the identification of novel diagnostic markers in prostate cancer. Expert Rev Mol Diagn 2015;15:1211-24. [Crossref] [PubMed]
- Lucarelli G, Ditonno P, Bettocchi C, et al. Serum sarcosine is a risk factor for progression and survival in patients with metastatic castration-resistant prostate cancer. Future Oncol 2013;9:899-907. [Crossref] [PubMed]
- Ochocki JD, Khare S, Hess M, et al. Arginase 2 Suppresses Renal Carcinoma Progression via Biosynthetic Cofactor Pyridoxal Phosphate Depletion and Increased Polyamine Toxicity. Cell Metab 2018;27:1263-80.e6. [Crossref] [PubMed]
- Huang HY, Wu WR, Wang YH, et al. ASS1 as a novel tumor suppressor gene in myxofibrosarcomas: aberrant loss via epigenetic DNA methylation confers aggressive phenotypes, negative prognostic impact, and therapeutic relevance. Clin Cancer Res 2013;19:2861-72. [Crossref] [PubMed]
- Kobayashi E, Masuda M, Nakayama R, et al. Reduced argininosuccinate synthetase is a predictive biomarker for the development of pulmonary metastasis in patients with osteosarcoma. Mol Cancer Ther 2010;9:535-44. [Crossref] [PubMed]
- Rabinovich S, Adler L, Yizhak K, et al. Diversion of aspartate in ASS1-deficient tumors fosters de novo pyrimidine synthesis. Nature 2015;527:379-83. [Crossref] [PubMed]


