Editorial on “The genomic landscape of TERT promoter wildtype-IDH wildtype glioblastoma”
In 2016, with the publication of the novel guidelines for the classification of the central nervous system (CNS) tumours by the World Health Organization (WHO) (1), there was a paradigm shift in the classical diagnosis, mainly based in microscopy, by the integration of molecular markers in the novel stratification. Due to the intrinsic limitations of the collected CNS study material and the majority of the conservative procedures that ends in scarce material, and sometimes of difficult histology, this added value of the molecular data introduced a significant advance in tumour homogenous classification. The genetic classification is based on canonical events that have been collected throughout the recent years (2-5). Regarding the pivotal events that are used to stratify glioblastoma (GBM), we have the mutations in the isocitrate dehydrogenase 1 and 2 (IDH) genes (2,5) and in the promoter of the telomerase gene (TERTp) (5-10). These alterations are used to aid in the diagnosis of up to 80% of GBM and can cluster patients with distinct prognostic features (5). With the molecular subgroups there is also an association for a particular mechanism used for telomere maintenance. Telomere maintenance can rely in mechanisms that are dependent in telomerase re-activation or re-expression, or in a telomerase non-dependent mechanism, the so-called alternative mechanism for telomere maintenance (ALT) (11). In a telomerase re-activation setting, TERTp mutations are so far recognized as the most frequent event and have been reported in several human cancers (6,8,12-15). In an ALT mechanism setting, the Alpha Thalassemia/Mental Retardation Syndrome X-Linked (ATRX) and Death Domain Associated Protein (DAXX) gene mutations were documented as the most frequently altered genes and were, until the study by Diplas et al. (16), the only known genes to be directly involved in ALT mechanism promotion. TERTp mutations are present in about 70% of IDH-wildtype GBM, associated with older patients that present a poorer prognosis with a shorter survival; this subgroup represents up to 90% of GBM (1,5). The remaining are IDH-mutant GBM, and present ATRX mutations in 70% of the cases (with ALT phenotype); this subgroup is composed of younger patients and presents a better outcome with increase overall survival (OS) when compared with the previous (1,4,5,8). In the study by Diplas et al. (16), the authors set to determine the genetic landscape of TERTpWT-IDHWT GBM. For this purpose, they identified a cohort of 16.9% of TERTpWT-IDHWT GBM from a series of 260 GBM that had been previously studied (5); of these, when 1p/19q co-deletion status was available it was negative, in this way this subset lack the three main glioma markers (TERTpWT-IDHWT-1p/19qWT)—triple negative tumours (17). The whole exome sequencing of this subgroup presented recurrent mutated genes in classical pathways as the RTK/RAS/PI3K (88%), P53 (40%), and RB (24%) pathways, as well as, copy number variations in PDGFRA (8%), MDM2 and MDM4 (12%) and CDKN2B (12%) genes. Analysis for glioma-associated driver alterations identified mutations in classical associated genes as PTEN (32%), NF1 (24%), EGFR (28%), TP53 (24%), ATRX (20%), and BRAF (20%), and in two novel candidate drivers that were not previously associated with GBM, SMARCAL1 (16%) and PPM1D (8%). The results obtained revealed several interesting findings. There was an enrichment of BRAF (V600E) mutations (20%) in the TERTpWT-IDHWT GBM, whereas BRAF alterations that were previously associated to low-grade pediatric gliomas, being rare in adult gliomas (18,19), had a high representation in this subgroup; with 80% of the patients being less or equal than 30 years old. The second novelty with this study was the detection of mutations in SMARCAL1 and PPM1D as novel gliomagenesis genes. The SMARCAL1 gene, that stands for the SWI/SNF Related, Matrix Associated, Actin Dependent Regulator Of Chromatin, Subfamily A Like 1 protein was the main focus of the study since it encodes an adenosine triphosphate (ATP)-dependent annealing helicase responsible for rewinding of Replication Protein A (RPA)-bound DNA at stalled replication forks for resolving telomere-associated replication stress (5); such similarity with ATRX function, also a member of the SWI/SNF family of chromatin remodelers, made it immediately an attractive candidate (16,20,21). All these facts led the authors to expand the cohort of TERTpWT-IDHWT GBM, and in 21% of these cases, SMARCAL1 mutations were detected. Given the similarities with ATRX, it was determined if SMARCAL1 inactivation was comparable to ATRX loss-of-function, and ALT initiation. The authors performed a battery of assays demonstrating classical ALT phenotypic features (16,20,21), such as the presence of ultrabright telomeric foci and C-circles and establishing a novel link of a gene to ALT development. Additionally, SMARCAL1-mutant GBM were mutually exclusive of GBM with ATRX loss of expression, reinforcing the independent contribution of each gene for the ALT mechanism. However, the authors noted that still other mechanisms should be involved because 61.5% of TERTpWT-IDHWT GBM remained without a telomere maintenance mechanism. To decipher further mechanisms, it was taken a whole genome sequencing approach in 8 TERTpWT-IDHWT GBM and structural variants (SV) of TERT were identified in 75% of the cases. This structural alteration consisted in half of the cases in translocations to other chromosomes and in the remaining cases in inversions within the same chromosome. In the expanded analysis of the TERTpWT-IDHWT GBM for TERT SV identification, the authors used break-apart FISH probes and found that half of the cases presented TERT structural rearrangements. Functionally, TERT-rearranged GBM expressed significantly higher levels of telomerase in comparison with the ALT-positive (ATRX- or SMARCAL1-mutated GBM) but with no striking differences in comparison with TERTp mutated GBM.
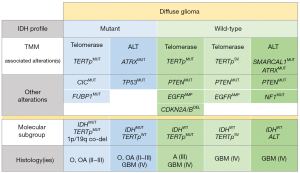
All the previous data together allowed the creation of new genetic subgroups of TERTpWT-IDHWT GBM (Figure 1). Within the previous IDHWT-ALT subgroup we are now aware that besides ATRX mutations (with absence of IDH and TP53 mutations), there was a fraction of cases that presented SMARCAL1 mutations (38.5%), a novel mechanism with phenotypic markers compatible with ALT. The SMARCAL1-mutant GBM contrary to the ATRX-mutated GBM often presented mutations in TP53 as well as in PTEN and NF1; such co-occurrences may be necessary for gliomagenesis. In addition to the subgroups with ALT, we have the IDHWT GBM subgroup that rely on telomerase re-expression due to TERT SVs. Altogether, this novel genetic events account for more than 80% of the TERTpWT-IDHWT GBM. In the study by Diplas et al. (16), it was also evaluated how this subgroup genetic integration impacts patients’ survival. Both IDHWT GBM, either with ALT or with TERT SVs exhibited the poorest OS (14.9 and 19.7 months, respectively), similarly to the IDHWT GBM with TERTp mutations (OS: 14.7 months); contrasting with the better outcome of GBM patients with IDH mutations and absence of TERTp alterations (OS: 37.1 months).
Apart from the definition of these novel subgroups of TERTpWT-IDHWT GBM, there was also a novel finding, the discovery of a new gene associated to ALT. The authors created a set of experiments to elucidate SMARCAL1 contribution for this process. The first approach was to start with cancer cell lines that presented mutations in this gene and to evaluate if these cell lines recapitulated the findings detected in the human GBM samples. Both SMARCAL1-mutant cell lines had abolished expression of the SMARCAL1 protein and maintained intact expression of ATRX and DAXX. Still, ALT phenotypical markers were present and included ALT-associated promyelocytic leukemia (PML) bodies (APBs), DNA C-circles, and the classical ultrabright telomere DNA foci by FISH (22); exogenous expression of SMARCAL1 was able to restore, or partially restore, the previous changes. The second approach went the opposite way and was the evaluation of GBM cell lines after CRISPR/Cas9 gene removal of the SMARCAL1 gene. Under this approach, isogenic SMARCAL1−/− GBM cell lines were assessed for the ALT markers, and it was observed that there was a significant increase of C-circles, ultrabright telomere foci formation and APBs presence; this increase was more evident when the alterations that targeted SMARCAL1 helicase domains, pointing its domain important role.
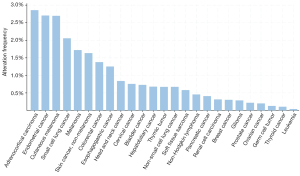
Overall, in the study by Diplas et al. (16), two novel subgroups of TERTpWT-IDHWT GBM were identified. They represent two genetically defined GBM subgroups, IDHWT-ALT and IDHWT-TERTSV that present similarities with the established IDHMUT and TERTpMUT, both subgroups relying in novel genetic alterations result in ALT-mediated or telomerase-mediated mechanisms for telomere maintenance with novel alterations (Figure 1). Within the gliomas, SMARCAL1 mutations seem to be rare in lower-grade gliomas (WHO grade II–III) and only present in high grade gliomas (WHO grade 4); also, TERT SV were only detected in GBM (WHO grade 4). The detection of a high frequency of BRAF mutations was a novelty that opens a new therapeutic option for this subgroup of patients younger than 30 years old, since this is an alteration that is drug-targetable. It remains now to be understood what is the true expression of this novel gene associated to ALT, the SMARCAL1 and its prevalence within cancer. In a short overview of the cancer genomic data accessed in cBioPortal (23,24) for cancer genomics (Figure 2) we detected that, besides GBM, several cancers present mutations in SMARCAL1 and despite the prevalence of these alterations to be inferior to 3% it may be useful to select patients with particular clinicopathological features.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned and reviewed by the Section Editor Duo Liu (Harbin Medical University Cancer Hospital, Harbin Medical University, Harbin, China).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.08.06). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Louis DN, Ohgaki H, Wiestler OD, et al. WHO classification of tumours of the central nervous system. World Health Organization classification of tumours. Lyon: International Agency For Research On Cancer (IARC), 2016.
- Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360:765-73. [Crossref] [PubMed]
- Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321:1807-12. [Crossref] [PubMed]
- Jiao Y, Killela PJ, Reitman ZJ, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget 2012;3:709-22. [Crossref] [PubMed]
- Killela PJ, Pirozzi CJ, Healy P, et al. Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget 2014;5:1515-25. [Crossref] [PubMed]
- Vinagre J, Almeida A, Populo H, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun 2013;4:2185. [Crossref] [PubMed]
- Batista R, Cruvinel-Carloni A, Vinagre J, et al. The prognostic impact of TERT promoter mutations in glioblastomas is modified by the rs2853669 single nucleotide polymorphism. Int J Cancer 2016;139:414-23. [Crossref] [PubMed]
- Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A 2013;110:6021-6. [Crossref] [PubMed]
- Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell 2013;155:462-77. [Crossref] [PubMed]
- Labussiere M, Boisselier B, Mokhtari K, et al. Combined analysis of TERT, EGFR, and IDH status defines distinct prognostic glioblastoma classes. Neurology 2014;83:1200-6. [Crossref] [PubMed]
- Gaspar TB, Sa A, Lopes JM, et al. Telomere Maintenance Mechanisms in Cancer. Genes (Basel) 2018;9: [Crossref] [PubMed]
- Huang FW, Hodis E, Xu MJ, et al. Highly recurrent TERT promoter mutations in human melanoma. Science 2013;339:957-9. [Crossref] [PubMed]
- Horn S, Figl A, Rachakonda PS, et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013;339:959-61. [Crossref] [PubMed]
- Vinagre J, Pinto V, Celestino R, et al. Telomerase promoter mutations in cancer: an emerging molecular biomarker? Virchows Arch 2014;465:119-33. [Crossref] [PubMed]
- Vinagre J, Nabais J, Pinheiro J, et al. TERT promoter mutations in pancreatic endocrine tumours are rare and mainly found in tumours from patients with hereditary syndromes. Sci Rep 2016;6:29714. [Crossref] [PubMed]
- Diplas BH, He X, Brosnan-Cashman JA, et al. The genomic landscape of TERT promoter wildtype-IDH wildtype glioblastoma. Nat Commun 2018;9:2087. [Crossref] [PubMed]
- Eckel-Passow JE, Lachance DH, Molinaro AM, et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med 2015;372:2499-508. [Crossref] [PubMed]
- Basto D, Trovisco V, Lopes JM, et al. Mutation analysis of B-RAF gene in human gliomas. Acta Neuropathol 2005;109:207-10. [Crossref] [PubMed]
- Horbinski C. To BRAF or not to BRAF: is that even a question anymore? J Neuropathol Exp Neurol 2013;72:2-7. [Crossref] [PubMed]
- Heaphy CM, de Wilde RF, Jiao Y, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science 2011;333:425. [Crossref] [PubMed]
- Jiao Y, Shi C, Edil BH, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 2011;331:1199-203. [Crossref] [PubMed]
- Amorim JP, Santos G, Vinagre J, et al. The Role of ATRX in the Alternative Lengthening of Telomeres (ALT) Phenotype. Genes (Basel) 2016;7: [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]