
EGFR exon 19 deletion switch and development of p.L792Q mutation as a new resistance mechanism to osimertinib: a case report and literature review
Introduction
The identification of genomic alterations harbored in non-small cell lung cancer (NSCLC) patients, in particular in epidermal growth factor receptor (EGFR) gene, led to a milestone in the improvement of treatment choices with first (gefitinib and erlotinib) and second (afatinib) generation tyrosine kinase inhibitors (TKIs) (1-4). However, the appearance of EGFR p.T790M exon 20 point mutation, represents the most common resistance mechanism for the first and second generation TKIs; afterwards this evidence, a third generation EGFR TKIs, such as osimertinib, have been introduced to treat these patients (5). Therefore, new mechanisms of resistance also against third generation TKI were recently described (1-5); the first of these identified mechanisms was the EGFR exon 20 p.C797S point mutation (6). Recently, we assessed the mutational status of a never-smoker male 68-year-old with lung adenocarcinoma (ADC) diagnosed in stage IV, harboring an exon 19 EGFR p.E746_A750del treated with gefitinib for 18 months. At the progression to first line treatment, the oncologist requested a liquid biopsy analysis showing the appearance of a resistant point mutation in EGFR exon 20 (p.T790M) in concomitance with the initial EGFR p.E746_A750del. Thus, the patient started a therapy with osimertinib. After 9 months of treatment with osimertinib, a total body computed tomography (CT) shows the development of a progression and the oncologist requested another liquid biopsy analysis. The obtained results showed, in absence of a previously detected EGFR p.E746_A750del and p.T790M mutation, the presence of a new exon 19 deletion (p.L747_A750>P) in concomitance with an uncommon EGFR exon 20 point mutation (p.L792Q). Here we aim to describe and discuss this case in the landscape of literature data.
Case presentation
A 68-year-old man, with an history of smoking, showed dyspnea and abdominal pain in the liver region. Thus, a total body CT was performed and showed a mass (40 mm × 27 mm) in the right lung and two additional masses (26–71 mm) in the liver. On December 2013 a CT-guided fine needle aspiration (FNA) on a liver lesion was performed. The microscopic analysis showed poorly differentiated neoplastic cells and, according to immunophenotypical features (nuclear positivity for TTF-1), a diagnosis of NSCLC favor ADC was made. At the time of the diagnosis the patient presented liver metastasis, so the tumor was classified in stage IV.
The previous international guidelines [2013] from the College of American Pathologists (CAP), International Association for the Study of Lung Cancer (IASLC), and Association for Molecular Pathology (AMP) suggested that in case of newly diagnosed IIIB/IV NSCLC molecular tests were recommended to better define the therapeutic strategy (7). Consistent with these, on January 2014 we performed a fragment analysis assay (for exon 19 deletions) and a TaqMan based real-time polymerase chain reaction (RT-PCR—for exon 21 point mutations), following a previously validated protocol (8,9). The result was a 15 base pairs deletion in EGFR exon 19, confirmed by a Sanger sequencing and re-analyzed by a next generation sequencing (NGS) approach, which showed a p.E746_A750del. Besides on this result, the patient started a treatment with gefitinib. This latter had been carried out from January 2014 to June 2015. Due to progression disease with the increase of the previously described lesions and appearance of new lesions, on August 2015 the patient started a treatment with 4 cycles of carboplatin-paclitaxel. In October 2015 was requested at our Institution the assessment, on circulating tumor DNA (ctDNA) derived from liquid biopsy, of exon 20 EGFR resistance mutation (p.T790M) in order to administrate a third generation TKI in the osimertinib expanded access study (10). The EGFR mutational status assessment was performed by the previously validated SiRe® NGS panel on Ion Torrent Personal Genome Machine (PGM, Thermofisher, Waltham, Massachusetts, USA) (11). The patient showed the resistance point mutation in EGFR exon 20 (p.T790M) in concomitance with the exon 19 deletion (p.E746_A750del) and underwent a treatment with osimertinib. In May 2017 the patient underwent another liquid biopsy analysis due to the re-presentation of abdominal pain and asthenia. We identified a mutation switch respect to the initial EGFR exon 19 deletion (p.L747_A750>P), and the uncommon point mutation in EGFR exon 20 (p.L792Q), without the evidence of p.T790M (Figure 1).
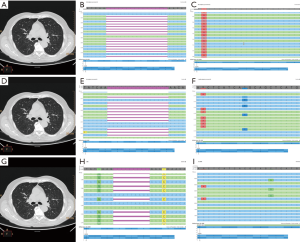
Discussion
Treatment with first or second generation TKIs leads to the development of resistance mutations (e.g., p.T790M) which make consequently neoplastic cells responsive to the third generation TKIs (e.g., osimertinib) (5). In these patients, the ctDNA analysis could be a valid option to investigate the development of others EGFR gene mutations (12,13). However, even under treatment with third generation TKI, neoplastic cells can develop several resistance mechanisms (Tables 1,2) (6,14-23). The first described is the point mutation p.C797S in EGFR exon 20 (6). In the study from Thress et al., six patients showed, in plasma samples, the p.C797S in association with the an exon 19 deletion and the p.T790M mutation (6). Other known resistance mutations involve EGFR codon 796 (14,15). In fact, Zheng et al., by using a NGS approach, detected a p.G796D mutation whose mechanism is a conformational interference in the interaction between osimertinib and EGFR kinase domain (14). Also this patients, as reported in our case, lost the initial sensitizing mutation (p.L858R) and p.T790M after progression to osimertinib (14). Ou et al. reported in a single patient the development of different resistance mutations (p.G796S/R, p.L792F/H, p.C797S/G and V802F) all in trans with each other and in cis with the p.T790M (15). As in our case, Chen et al. identified on ctDNA derived from three patients (n=2 plasma samples and n=1 pleural effusion), acquired resistance mutations on EGFR in codon 792 (L792F, L792Y and L792H) (16). Interestingly, all these alterations were in cis with the p.T790M and in trans with the p.C797S (16). In a case by Bersanelli et al. a novel p.L718Q EGFR point mutation was evaluated as a resistance mechanism against osimertinib, without the evidence of p.C797S or other known EGFR-independent mechanisms of resistance (17). Oztan et al. described other two cases (in tissue and blood samples respectively) in which an EGFR p.G724S was detected, with or without the p.T790M (18). Alternative mechanism of acquired resistance to osimertinib involved the BRAF pathway, as shown by Ho et al. (19). In this study, by using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), they identified, on tissue sample, a BRAF p.V600E mutation (19); in particular, the first EGFR mutation (p.L858R) was lost or not yet detectable, after treatment with gefitinib and erlotinib, and the patient acquired a concomitant exon 19 deletion with p.T790M that can be detected also after progression to osimertinib in concomitance with the BRAF p.V600E (19). Furthermore, Planchard et al. described other mechanisms of resistance, in two different patients who underwent a therapy with a third generation TKI (20); the authors identified by comparative genomic hybridization (CGH) and fluorescent in situ hybridization (FISH), HER2 and MET amplification (20). In both cases these alterations were correlated with the loss of the EGFR p.T790M mutation (20). In another experience, Ou et al. also identified a MET amplification developed after treatment with osimertinib, and in accordance with Planchard et al., no evidence of this alteration was found before treatment (21). Another recent experience, reported by Knebel et al. showed, by using digital droplet PCR (ddPCR), an amplification of the EGFR exon 19 deleted allele without the evidence of other known resistance mechanisms to osimertinib (22). Ham et al. in two different cases, harboring two different EGFR sensitizing mutation, identified a transition from adenocarcinoma to small cell carcinoma after osimertinib with the concomitant loss of the p.T790M mutation (23). As in our case, Ho et al. showed the switch of the initial sensitizing EGFR mutation (p.L858R) with another mutation (exon 19 deletion) (19). Instead, different from our experience, their case retain the p.T790M mutation with the acquisition of a concomitant BRAF p.V600E (19). The heterogeneity of EGFR mutational status has been described in literature, and also the possibility of arising of different clones after target treatment (24,25). Moreover, the development of an osimertinib resistance mutation (e.g., EGFR p.C797S) with the concomitant loss of p.T790M and persistence of an EGFR sensitizing mutation allows the re-treatment with a first or second generation TKI, as recently reported by Chic et al. (26).
Table 1
| First author | EGFR initial mutation (persistence at the progression to osimertinib) | First TKI adopted | Resistance mechanism described | Persistence at the progression to osimertinib of p.T790 | Methodology adopted |
|---|---|---|---|---|---|
| Thress et al. (6) | Exon 19del (Y) | NR | p.C797S | Y | NGS |
| Zheng et al. (14) | p.L858R (N) | Gefitinib | p.G796D | N | NGS |
| Ou et al. (15) | p.L858R (Y) | Erlotinib | p.G796S/R, p.L792F/H, p.C797S/G, p.V802F | Y | HC NGS |
| Chen et al. (16) | Exon 19del (Y) | Gefitinib | p.L792H/F, p.C797S/G/N, p.L718Q | Y | Targeted NGS |
| Exon 19del (Y) | Gefitinib | p.L792H/F/Y, p.C797S | Y | ||
| Exon 19del (Y) | Gefitinib | p.L792F, p.C797S, p.P794S | Y | ||
| Bersanelli et al. (17) | p.L858R (Y) | Gefitinib | p.L718Q | Y | NGS |
| Oztan et al. (18) | Exon 19del (Y) | Erlotinib | p.G724S | Y | CGP |
| Exon 19del (Y) | Erlotinib | p.G724S | N | ||
| Pisapia, et al. (present study) | Exon 19del (N) | Gefitinib | p.L792Q | N | NGS |
TKI, tyrosine kinase inhibitor; HC NGS, hybrid capture based next generation sequencing; CGP, comprehensive genome profiling.
Table 2
| First author | EGFR initial mutation (persistence at the progression to osimertinib) | First TKI adopted | Resistance mechanism described | Persistence at the progression to osimertinib of p.T790 | Methodology adopted |
|---|---|---|---|---|---|
| Ho et al. (19) | p.L858R (N) | Gefitinib and erlotinib | p.V600E (BRAF) | Y | MALDI-TOF MS |
| Planchard et al. (20) | p.E746_A750del (Y) | Gefitinib | HER2 amplification | N | CGH, FISH |
| p.L858R (Y) | Erlotinib | MET amplification | N | ||
| Ou et al. (21) | Exon 19del (NR) | Erlotinib | MET amplification | Y | CGP |
| Knebel et al. (22) | Exon 19del (Y) | Erlotinib | EGFR amplification | Y | ddPCR |
| Ham et al. (23) | p. L858R (Y) | Erlotinib | SCLC transformation | N | NGS |
| Exon 19del (Y) | Erlotinib | SCLC transformation | N |
CGH, comparative genomic hybridization; CGP, comprehensive genome profiling; ddPCR, digital droplet polymerase chain reaction; FISH, fluorescent in situ hybridization; HC NGS, hybrid capture based next generation sequencing; MALDI-TOF MS, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; NGS, next generation sequencing; NR, not reported; SCLC, small cell lung cancer; Y, yes; N, no.
In conclusion, we report an uncommon mechanism of resistance to osimertinib, identified with a validated NGS approach, different from the classic point mutation p.C797S, which involved, in addition to the uncommon point mutation p.L792Q, the switch from the original sensitizing EGFR deletion (from p.E746_A750del to p.L747_A750>P) and the loss of p.T790M mutation.
Acknowledgments
Funding: None.
Footnote
Provenance and peer review: The article was commissioned by the editorial office, Translational Cancer Research for the series “Targeted Therapy and Non-Small Cell Lung Cancer: A New Era?”. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.09.13). The series “Targeted Therapy and Non-Small Cell Lung Cancer: A New Era?” was commissioned by the editorial office without any funding or sponsorship. UM served as the unpaid Guest Editor of the series. The authors have no other conflicts of interest to declare.
Ethical statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient for publication of this manuscript and any accompanying images.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Karachaliou N, Mayo-de las Casas C, Queralt C, et al. Association of EGFR L858R mutation in circulating free DNA with survival in the EURTAC Trial. JAMA Oncol 2015;1:149-57. [Crossref] [PubMed]
- Yang JC, Wu YL, Schuler M, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol 2015;16:141-51. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560-2. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med 2013;137:828-60. [Crossref] [PubMed]
- Malapelle U, Russo S, Pepe F, et al. EGFR mutation detection by microfluidic technology: a validation study. J Clin Pathol 2013;66:982-4. [Crossref] [PubMed]
- Malapelle U, Bellevicine C, De Luca C, et al. EGFR mutations detected on cytology samples by a centralized laboratory reliably predict response to gefitinib in non-small cell lung carcinoma patients. Cancer Cytopathol 2013;121:552-60. [Crossref] [PubMed]
- De Marinis F, Chul CB, Kim DW, et al. ASTRIS: A real world treatment study of osimertinib in patients (pts) with EGFR T790M positive non-small cell lung cancer (NSCLC). J Clin Oncol 2017;35:9036. [Crossref]
- Malapelle U, Mayo de-Las-Casas C, Rocco D, et al. Development of a gene panel for next- generation sequencing of clinically relevant mutations in cell-free DNA from cancer patients. Br J Cancer 2017;116:802-10. [Crossref] [PubMed]
- Malapelle U, Pisapia P, Rocco D, et al. Next generation sequencing techniques in liquid biopsy: focus on non-small cell lung cancer patients. Transl Lung Cancer Res 2016;5:505-10. [Crossref] [PubMed]
- Pisapia P, Pepe F, Smeraglio R, et al. Cell free DNA analysis by SiRe® next generation sequencing panel in non small cell lung cancer patients: focus on basal setting. J Thorac Dis 2017;9:S1383-90. [Crossref] [PubMed]
- Zheng D, Hu M, Bai Y, et al. EGFR G796D mutation mediates resistance to osimertinib. Oncotarget 2017;8:49671-9. [PubMed]
- Ou SI, Cui J, Schrock AB, et al. Emergence of novel and dominant acquired EGFR solvent-front mutations at Gly796 (G796S/R) together with C797S/R and L792F/H mutations in one EGFR (L858R/T790M) NSCLC patient who progressed on osimertinib. Lung Cancer 2017;108:228-31. [Crossref] [PubMed]
- Chen K, Zhou F, Shen W, et al. Novel Mutations on EGFR Leu792 Potentially Correlate to Acquired Resistance to Osimertinib in Advanced NSCLC. J Thorac Oncol 2017;12:e65-8. [Crossref] [PubMed]
- Bersanelli M, Minari R, Bordi P, et al. L718Q Mutation as New Mechanism of Acquired Resistance to AZD9291 in EGFR-Mutated NSCLC. J Thorac Oncol 2016;11:e121-3. [Crossref] [PubMed]
- Oztan A, Fischer S, Schrock AB, et al. Emergence of EGFR G724S mutation in EGFR-mutant lung adenocarcinoma post progression on osimertinib. Lung Cancer 2017;111:84-7. [Crossref] [PubMed]
- Ho CC, Liao WY, Lin CA, et al. Acquired BRAF V600E Mutation as Resistant Mechanism after Treatment with Osimertinib. J Thorac Oncol 2017;12:567-72. [Crossref] [PubMed]
- Planchard D, Loriot Y, André F, et al. EGFR-independent mechanisms of acquired resistance to AZD9291 in EGFR T790M-positive NSCLC patients. Ann Oncol 2015;26:2073-8. [Crossref] [PubMed]
- Ou SI, Agarwal N, Ali SM. High MET amplification level as a resistance mechanism to osimertinib (AZD9291) in a patient that symptomatically responded to crizotinib treatment post-osimertinib progression. Lung Cancer 2016;98:59-61. [Crossref] [PubMed]
- Knebel FH, Bettoni F, Shimada AK, et al. Sequential liquid biopsies reveal dynamic alterations of EGFR driver mutations and indicate EGFR amplification as a new mechanism of resistance to osimertinib in NSCLC. Lung Cancer 2017;108:238-41. [Crossref] [PubMed]
- Ham JS, Kim S, Kim HK, et al. Two Cases of Small Cell Lung Cancer Transformation from EGFR Mutant Adenocarcinoma During AZD9291 Treatment. J Thorac Oncol 2016;11:e1-4. [Crossref] [PubMed]
- Hata A, Yoshioka H, Fujita S, et al. Complex mutations in the epidermal growth factor receptor gene in non-small cell lung cancer. J Thorac Oncol 2010;5:1524-8. [Crossref] [PubMed]
- Remon J, Majem M. EGFR mutation heterogeneity and mixed response to EGFR tyrosine kinase inhibitors of non small cell lung cancer: a clue to overcoming resistance. Transl Lung Cancer Res 2013;2:445-8. [PubMed]
- Chic N, Mayo-de-Las-Casas C, Reguart N. Successful Treatment with Gefitinib in Advanced Non-Small Cell Lung Cancer after Acquired Resistance to Osimertinib. J Thorac Oncol 2017;12:e78-80. [Crossref] [PubMed]


