
miR-24-3p promotes cell migration and invasion by targeting TEL2 in nasopharyngeal carcinoma
Introduction
Nasopharyngeal carcinoma (NPC) is a malignancy that occurs in most of the world but has an especially high incidence in southern China, Southeast Asia and North Africa (1). Additionally, NPC has the highest metastasis rate, of which 19.9% and 74.5% of patients show distant metastasis and regional lymph node metastasis, respectively (2-4). In recent years, some progress has been made in the treatment of the distant metastasis of NPC; however, the underlying mechanism of NPC metastasis remains unclear.
TEL2 is a member of the ETS family, which plays an imperative role in the development of normal hematopoiesis (5) and tumor formation. All ETS factors share highly conserved DNA binding domains that interact specifically with DNA sequences (6). As transcription factors, they can regulate the expression of target genes in various signaling pathways to influence the growth, invasion and metastasis of cancer. TEL2 binds to DNA through the ETS domain and interacts with itself or TEL1 through the tip domain (7). Thus, TEL2 is a multifunctional transcription factor that regulates the expression of target genes in signaling pathways to influence the development of tumors, cell differentiation, apoptosis, and migration as well as invasion, and metastasis (8,9). In previous studies, we identified that TEL2 is an important regulatory molecule for NPC metastasis by gene microarray technology (10). However, it is unknown how TEL2 is downregulated in highly metastatic NPC cells.
MicroRNAs (miRNAs), a key component of the noncoding RNA family, play a significant role in the initiation and progression of cancer (11). miRNAs typically target multiple mRNAs and regulate the expression of genes by inhibiting the translation or degradation of target mRNAs. There are several studies showing that miR-24 is downregulated in osteosarcoma (12), gastric cancer (13), and bladder cancer (14,15), in addition to being upregulated in hepatocellular carcinoma (16,17) and breast cancer (18,19). Our previous study also showed that TEL2 is downregulated in highly metastatic NPC cells (10) and that the overexpression of TEL2 could inhibit the migration, invasion and metastasis of NPC. miR-24-3p has been shown to act as an oncogene or tumor suppressor in certain cancers (20). For example, as an oncogenic role, we not only reported that miR-24-3p is downregulated in colorectal cancer (CRC) and is associated with advanced disease (21), but it is also involved in pathogenesis that may serve as a potential diagnostic target. By contrast, it can play a suppressive role via downregulating LPAATb expression (22). However, recent miR-24-3p- and NPC-related studies have primarily focused on how miR-24-3p changes NPC radio-sensitivity by directly regulating Jab1/CSN5 (23). Furthermore, it also contributes to tumor pathogenesis via FGF11 repression by modulating T-cell function in NPC, forming a unique intercellular mechanism inside the human body (24,25). However, in this report, we demonstrated that the upregulation of miR-24-3p leads to TEL2 mRNA and protein downregulation. This new miR-24-3p/TEL2 pathway may provide a novel therapeutic direction for the treatment of NPC metastasis.
Methods
Cell lines and reagents
Cell lines (NP69, 6-10B, 5-8F, HONE1, S26, S18, and SUNE1) were provided by the group of Tiebang Kang (Sun Yat-sen University Cancer Center, Guangzhou, China). Cell lines (6-10B, S26, and CNE1) were cultured in DMEM (Gibco; Catalog number: 11965-092) supplemented with 10% fetal bovine serum (Gibco; Catalog number: 10270-106), and all cells were grown in a humidified incubator with the conditions of 37 °C and 5% CO2, and the cell culture was maintained for no more than 6 months.
Clinical samples and study approval
A total of 116 NPC samples were obtained in this experiment. The case data showed that these patients are between 6 and 52 years of age. The tissue of 116 patients with NPC was collected, and the mRNA expression levels of TEL2 and miR-24-3p were detected. Written informed consent was obtained from all patients before sample collection, and this study was approved by the Institutional Review Board of the Third Affiliated Hospital of Nanchang University (No. CAN20144895).
Transient transfection of miRNA mimics and inhibitors
The miRNA mimics (miR-24-3p), miRNA control, and miRNA inhibitors for miR-24-3p were purchased from Ribobio (Ribobio, Guangzhou, China). After seeding into 6-well plates, the cells were transfected with miRNA mimics at a final concentration of 100 nM using RNAiMAX Reagent (Invitrogen).
Plasmid transfection
Lipofectamine 2000 (Invitrogen) was used for transfection according to the manufacturer’s instructions. Briefly, NPC cells were inoculated with 2.5×105 cells per well in 6-well tissue culture dishes, and 5 g of plasmid DNA was transfected for 24 hours, followed by the use of indicator chemicals.
RNA extraction, reverse transcription, and quantitative real-time PCR
These procedures were performed as described previously (26,27). TRIzol reagent (Invitrogen) was used to isolate total RNA according to the manufacturer’s protocol (Thermo Fisher Scientific; Catalog number: 15596026). The first-strand cDNA was synthesized using the RevertAidTM First Strand cDNA Synthesis Kit (Thermo Scientific; Catalog number: K1621). Quantification of miR-24-3p was performed using the stem-loop real-time PCR miRNA kit (Ribobio, China), and small RNA U6 was used as an internal control. The primers for miR-24-3p and U6 were purchased from Ribobio. The primers used to amplify TEL2 and GAPDH were as follows: 5'-GGGCTTACCAGCAACTTCG-3' and 5'-TCTTGGCGTCCTTGTCTTCC-3'. The GAPDH primer sequences were as follows: 5'-ACAGTCAGCCGCATCTTCTT-3' and 5'-GACAAGCTTCCCGTTCTCAG-3'. Additionally, quantification reverse transcriptase-polymerase chain reaction (qRT-PCR) analysis was carried out on 3 independent RNA samples.
Western blotting
Western blotting was performed as described previously (28). The cells were collected and lysed with RIPA buffer [0.5% EDTA, 150 mm NaCl, 0.5% NP40, 50 mM Tris-HCl (pH 8.0)] and then were centrifuged at 12,000×g at 4 °C for 20 min. Next, 50 µg of total harvested protein was separated by 12% SDS-PAGE and then was separated in 8% sodium dodecyl sulfate-polyacrylamide gradient gels, followed by transfer onto polyvinylidene fluoride membranes (Bio-Rad Laboratories, Inc.). The membranes were blocked with 5% non-fat milk at room temperature for 2 hours and washed with TBST three times for 5 min each. The membranes were then later incubated with primary antibodies and horseradish peroxidase-conjugated secondary antibodies. After washing the PVDF membranes three times with TBST for 5 min each, they were finally detected using the ECL chemiluminescence system (Pierce, Rockford, USA).
Transwell assays
For the Transwell migration assay, 5.0×104 cells re-suspended in 300 µL of serum-free DMEM (Gibco; Catalog number: 11965-092) were added to cell culture inserts with 8 µm microporous filters without an extracellular matrix coating (Becton Dickinson Labware, Bedford, MA; Catalog number: 353097). DMEM containing 10% FBS (Gibco; Catalog number: 10270-106) was then added to the bottom chamber (Becton Dickinson Labware, Bedford, MA, Catalog number: 353504). After 24 hours of incubation, the cells on the surface of the filter paper were fixed, stained and examined under a microscope. The upper chamber cells were incubated with a cotton swab for 12 h, followed by incubation at 37 °C in a 5% CO2 incubator for 12 h. The cells were then stained with 95% ethanol for 15 min and crystal violet for 10 min. Under the microscope, 6 fields were evaluated randomly, and the number of migratory cells in the three random optical fields (×100 magnification) from the triplicate filter was averaged. In the Transwell invasion assay, the Transwell chamber containing Matrigel was purchased from BD (Cat: 354480). Next, 1×105 cells were suspended in 300 µL of serum-free DMEM and were added to the cell inserts. The following steps were performed as those for the migration assay described above. The number of cells that passed through the membrane was statistically analyzed using t-test.
Luciferase reporter assay
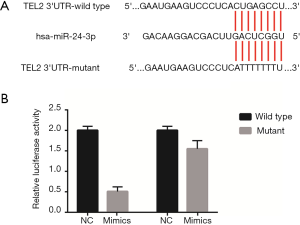
This process was carried out as described previously (26). The TEL2 gene 3' UTR wild-type or its mutant sequence (Figure 1) was linked to the luciferase reporter vector, and the recombinant plasmid pGL3 Report TEL2 3' UTR wild-type or mutant was constructed. The recombinant plasmid was co-transfected with miR-24-3p into target cells after transfection for 48 h according to the luciferase assay kit to detect firefly luciferase, and luciferase activity was calculated as the relative luminescence ratio, according to the ratio of the luciferase activity of the two groups.
Statistical analysis
Statistical analyses were performed using SPSS 22.0 software (IBM SPSS, Armonk, NY, USA). Two-tailed Student’s t-test was used to evaluate the differences between the two groups of data in all relevant experiments. The relationship between TEL2 and miR-24-3p expression was assessed using two-tailed Pearson’s correlation. Differences were statistically significant if P<0.01 or P<0.05 as indicated in the figure legends. All values from the in vitro assays are expressed as the means ± SD or the means ± SEM of at least three independent experiments or replicates.
Results
miR-24-3p negatively regulates the TEL2 mRNA and protein levels
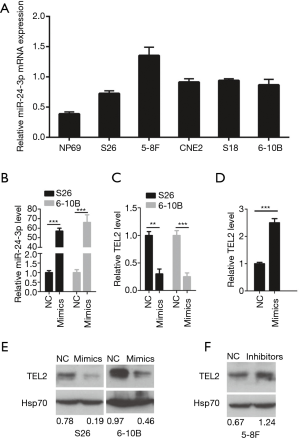
Using bioinformatics predictions (target scan) (29,30), we found that the 3' UTR of TEL2 (position 337-343) is a potential seeding site for miR-24-3p. Subsequently, we detected the expression of miR-24-3p mRNA in NP69 cells and five NPC cell lines. Compared with the NP69 cell line, the results showed that miR-24-3p is highly expressed in NPC cell lines, such as S26, S18, CNE2, 5-8F and 6-10B (Figure 2A). To explore the effects of miR-24-3p on the TEL2 mRNA and protein levels in NPC, we transfected miR-24-3p mimics into S26 and 6-10B cells. As shown in Figure 2B, qRT-PCR results showed that the expression of miR-24-3p was significantly upregulated in the miR-24-3p mimics transfection group in S26 and 6-10B cell lines compared with that in the NC control group. As expected, the expression of TEL2 was significantly downregulated in the miR-24-3p mimics group (Figure 2C) compared with that in the NC control group. By contrast, in 5-8F cells, TEL2 expression was significantly upregulated in the miR-24-3p inhibitors group compared with that in the control group (Figure 2D). Moreover, western blotting showed that the protein expression of TEL2 was decreased after transfecting with miR-24-3p mimics (Figure 2E) but increased when transfecting miR-24-3p inhibitors (Figure 2F). The above results suggested that miR-24-3p negatively regulated the TEL2 mRNA and protein levels.
miR-24-3p directly inhibits the expression of TEL2
Subsequently, we asked whether miR-24-3p could directly target TEL2, and the wild-type and mutant 3' UTR of TEL2 were cloned into a luciferase reporter vector (Figure 1A). miR-24-3p significantly inhibited the luciferase activity of pGL3-TEL2 3' UTR WT (Figure 1B), while the binding site containing the mutation abolished the effect of miR-24-3p. These results indicated that TEL2 is a direct target of miR-24-3p in NPC cells.
TEL2 is inversely correlated with miR-24-3p in NPC
To further investigate the correlation between TEL2 and miR-24-3p in NPC, 116 NPC patient samples were collected, total RNA was extracted for qRT-PCR, and the expression levels of TEL2 and miR-24-3p were measured. GAPDH and U6 were selected as controls for TEL2 and miR-24-3p, respectively. The results in Figure 3 clearly showed a significant negative correlation between TEL2 and miR-24-3p mRNA levels in NPC.

miR-24-3p promotes NPC cell migration and invasion by suppressing TEL2
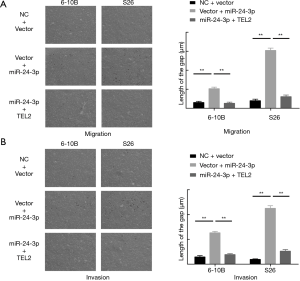
Given that TEL2 suppresses the migration, invasion and metastasis, we performed rescue assays. Transwell assays were additionally conducted to study the effect of cancer cell migration and invasion; as expected, miR-24-3p increased the abilities of NPC cell migration and invasion, but co-transfection of miR-24-3p and TEL2 can rescue miR-24-3p-mediated cell migration and invasion (Figure 4A,B). Collectively, our results showed that miR-24-3p promotes NPC cell migration and invasion by suppressing TEL2.
Discussion
In this report, we found that miR-24-3p expression is higher in NPC cell lines than in NP69 cells. We found that miR-24-3p had a negative regulatory effect on the mRNA and protein levels of TEL2 by targeting its 3' UTR. We also showed miR-24-3p could effectively promote migration and invasion by suppressing TEL2. Thus, the new axis of miR-24-3p/TEL2 may have a positive effect on the future treatment of NPC
Our previous findings indicated that TEL2 was low expressed in highly metastatic NPC cells and that the ectopic expression of TEL2 could inhibit the migration, invasion and metastasis of NPC (10). In this study, overexpression of miR-24-3p mimics in S26 and 6-10B cells significantly reduced the mRNA and protein levels of TEL2, while the inhibitor of miR-24-3p in S18 cells upregulated the mRNA and protein levels of TEL2. Our study revealed that miR-24-3p could be a direct negative regulator of TEL2.
Most of the research previously performed on TEL2 was focused on leukemia and hematopoiesis (5,31,32). In fact, previous studies have reported that TEL2 is a key player in NPC metastasis because it directly reduces the expression of SERPINE1 (10). However, more significantly, the high expression of SERPINE1 can promote the metastasis of NPC resulting from the low expression of TEL2. It was found that the high expression of TEL2 in patients with NPC showed a better prognosis than in patients with low expression of TEL2. These data demonstrated that TEL2 may have an important tumor-suppressive role in NPC.
miRNAs were considered to play an essential regulatory role in the invasion and metastasis of NPC (33). Several miRNAs targeting cancer regulatory molecules are involved in a complex signaling network in the tumor microenvironment (34), especially in metastasis and invasion (35). Although miRNAs are widely involved in the progress of cancer, its regulatory mechanisms have largely remained unexplored.
Conclusions
In conclusion, we confirm that miR-24-3p negatively regulates TEL2 directly. We also might be the first to systematically show that miR-24-3p plays a vital role in NPC metastasis—clarifying the pathogenesis of NPC to lay the foundation for future cancer diagnosis and treatment. This may also help clarify the occurrence and development of NPC, as well as its metastasis mechanism, to provide new molecular targets for the prevention and treatment of NPC. However, further research needs to be conducted in animal models to confirm whether the negative regulation of TEL2 by miR-24-3p can be clinically applied.
Acknowledgments
BF Xiao would like to thank Liang-Ke Liu who is at Tsinghua University, for her patience and consideration, which has given him great support in the duration of last year.
Funding: This work was supported by the National Science Foundation of China (Grant No. 81660449 to Y Sang) and the Jiangxi Provincial Natural Science Foundation of China (Grant No. 20161ACB21001, No. 20171BCD40026, both to Y Sang).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.10.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of the Third Affiliated Hospital of Nanchang University (No. CAN20144895) and written informed consent was obtained from all patients before sample collection.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bei JX, Jia WH, Zeng YX. Familial and large-scale case-control studies identify genes associated with nasopharyngeal carcinoma. Semin Cancer Biol 2012;22:96-106. [Crossref] [PubMed]
- Lee AW, Law SC, Foo W, et al. Nasopharyngeal carcinoma: local control by megavoltage irradiation. Br J Radiol 1993;66:528-36. [Crossref] [PubMed]
- Geara FB, Sanguineti G, Tucker SL, et al. Carcinoma of the nasopharynx treated by radiotherapy alone: determinants of distant metastasis and survival. Radiother Oncol 1997;43:53-61. [Crossref] [PubMed]
- Huang CJ, Leung SW, Lian SL, et al. Patterns of distant metastases in nasopharyngeal carcinoma. Kaohsiung J Med Sci 1996;12:229-34. [PubMed]
- Potter MD, Buijs A, Kreider B, et al. Identification and characterization of a new human ETS-family transcription factor, TEL2, that is expressed in hematopoietic tissues and can associate with TEL1/ETV6. Blood 2000;95:3341-8. [PubMed]
- Gu X, Shin BH, Akbarali Y, et al. Tel-2 is a novel transcriptional repressor related to the Ets factor Tel/ETV-6. J Biol Chem 2001;276:9421-36. [Crossref] [PubMed]
- Kawagoe H, Potter M, Ellis J, et al. TEL2, an ETS factor expressed in human leukemia, regulates monocytic differentiation of U937 Cells and blocks the inhibitory effect of TEL1 on ras-induced cellular transformation. Cancer Res 2004;64:6091-100. [Crossref] [PubMed]
- Chakrabarti R, Hwang J, Andres Blanco M, et al. Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nat Cell Biol 2012;14:1212-22. [Crossref] [PubMed]
- Kar A, Gutierrez-Hartmann A. Molecular mechanisms of ETS transcription factor-mediated tumorigenesis. Crit Rev Biochem Mol Biol 2013;48:522-43. [Crossref] [PubMed]
- Sang Y, Chen MY, Luo D, et al. TEL2 suppresses metastasis by down-regulating SERPINE1 in nasopharyngeal carcinoma. Oncotarget 2015;6:29240-53. [Crossref] [PubMed]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011;12:861-74. [Crossref] [PubMed]
- Liu Z, Liu Z, Zhang Y, et al. miR-24 represses metastasis of human osteosarcoma cells by targeting Ack1 via AKT/MMPs pathway. Biochem Biophys Res Commun 2017;486:211-7. [Crossref] [PubMed]
- Zhang H, Duan J, Qu Y, et al. Onco-miR-24 regulates cell growth and apoptosis by targeting BCL2L11 in gastric cancer. Protein Cell 2016;7:141-51. [Crossref] [PubMed]
- Yu G, Jia Z, Dou Z. miR-24-3p regulates bladder cancer cell proliferation, migration, invasion and autophagy by targeting DEDD. Oncol Rep 2017;37:1123-31. [Crossref] [PubMed]
- Zhang S, Zhang C, Liu W, et al. MicroRNA-24 upregulation inhibits proliferation, metastasis and induces apoptosis in bladder cancer cells by targeting CARMA3. Int J Oncol 2015;47:1351-60. [Crossref] [PubMed]
- Dong X, Ding W, Ye J, et al. MiR-24-3p enhances cell growth in hepatocellular carcinoma by targeting metallothionein 1M. Cell Biochem Funct 2016;34:491-6. [Crossref] [PubMed]
- Chen L, Luo L, Chen W, et al. MicroRNA-24 increases hepatocellular carcinoma cell metastasis and invasion by targeting p53: miR-24 targeted p53. Biomed Pharmacother 2016;84:1113-8. [Crossref] [PubMed]
- Cui S, Liao X, Ye C, et al. ING5 suppresses breast cancer progression and is regulated by miR-24. Mol Cancer 2017;16:89. [Crossref] [PubMed]
- Roscigno G, Puoti I, Giordano I, et al. MiR-24 induces chemotherapy resistance and hypoxic advantage in breast cancer. Oncotarget 2017;8:19507-21. [Crossref] [PubMed]
- Giglio S, Cirombella R, Amodeo R, et al. MicroRNA miR-24 promotes cell proliferation by targeting the CDKs inhibitors p27Kip1 and p16INK4a. J Cell Physiol 2013;228:2015-23. [Crossref] [PubMed]
- Gao Y, Liu Y, Du L, et al. Down-regulation of miR-24-3p in colorectal cancer is associated with malignant behavior. Med Oncol 2015;32:362. [Crossref] [PubMed]
- Song L, Yang J, Duan P, et al. MicroRNA-24 inhibits osteosarcoma cell proliferation both in vitro and in vivo by targeting LPAATbeta. Arch Biochem Biophys 2013;535:128-35. [Crossref] [PubMed]
- Wang S, Pan Y, Zhang R, et al. Hsa-miR-24-3p increases nasopharyngeal carcinoma radiosensitivity by targeting both the 3’ UTR and 5’ UTR of Jab1/CSN5. Oncogene 2016;35:6096-108. [Crossref] [PubMed]
- Ye SB, Li ZL, Luo DH, et al. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget 2014;5:5439-52. [Crossref] [PubMed]
- Ye SB, Zhang H, Cai TT, et al. Exosomal miR-24-3p impedes T-cell function by targeting FGF11 and serves as a potential prognostic biomarker for nasopharyngeal carcinoma. J Pathol 2016;240:329-40. [Crossref] [PubMed]
- Xu S, Feng Z, Zhang M, et al. hSSB1 binds and protects p21 from ubiquitin-mediated degradation and positively correlates with p21 in human hepatocellular carcinomas. Oncogene 2011;30:2219-29. [Crossref] [PubMed]
- Lv XB, Liu L, Cheng C, et al. SUN2 exerts tumor suppressor functions by suppressing the Warburg effect in lung cancer. Sci Rep 2015;5:17940. [Crossref] [PubMed]
- Sang Y, Wang L, Tang JJ, et al. Oncogenic roles of carbonic anhydrase IX in human nasopharyngeal carcinoma. Int J Clin Exp Pathol 2014;7:2942-9. [PubMed]
- Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92-105. [Crossref] [PubMed]
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4. [PubMed]
- Cardone M, Kandilci A, Carella C, et al. The novel ETS factor TEL2 cooperates with Myc in B lymphomagenesis. Mol Cell Biol 2005;25:2395-405. [Crossref] [PubMed]
- Carella C, Potter M, Bonten J, et al. The ETS factor TEL2 is a hematopoietic oncoprotein. Blood 2006;107:1124-32. [Crossref] [PubMed]
- Sengupta S, den Boon JA, Chen IH, et al. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc Natl Acad Sci U S A 2008;105:5874-8. [Crossref] [PubMed]
- Gutierrez NC, Sarasquete ME, Misiewicz-Krzeminska I, et al. Deregulation of microRNA expression in the different genetic subtypes of multiple myeloma and correlation with gene expression profiling. Leukemia 2010;24:629-37. [Crossref] [PubMed]
- Zhang Y, Yang P, Wang XF. Microenvironmental regulation of cancer metastasis by miRNAs. Trends Cell Biol 2014;24:153-60. [Crossref] [PubMed]


