
Critical role of KRAS mutation in pancreatic ductal adenocarcinoma
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal human cancers (1), with a 5-year survival rate of less than 7%, and a great proportion of patients die within 6 months after diagnosis. Previous reports have indicated that in 2017 more than 91,500 patients died of PDAC in the European Union (2). Various hereditary changes—for instance, genetic deletions, amplifications, translocations, frameshifts, inversions and substitutions—can be found in approximately 97% of PDAC cases (3). KRAS mutation is the most frequent mutation (more than 90%) (4) and the initiating genetic event for PDAC, and it is found in primary tumors, metastatic tumors and even in pancreatic intraepithelial neoplasia (PanIN), the earliest preneoplastic stage in PDAC progression (5).
As one of the four main driver genes (KRAS, TP53, CDKN2A and SMAD4) in PDAC, the KRAS gene, located at chromosome 12p12.1, is a member of the RAS gene family and encodes the KRAS protein (21 kDa), which has GTPase activity and thus binds GTP in the activated state and GDP in the deactivated state. Ras regulates cell proliferation, differentiation, and apoptosis by activating several signaling pathways, including the RAF/MEK/ERK, PI3K/AKT/mTOR, PLC/PKC, and RAL pathways (6). KRAS mutations are found at codon12, codon13, codon60 and codon61 in PDAC, and these mutations cause KRAS protein to remain activated in the absence of signal stimulation, resulting in an uncontrollable functional status. This review summarizes the current knowledge regarding the critical applications of KRAS mutation in PDAC.
Microenvironment
The tumor microenvironment, which is a dynamic network primarily comprising the matrix, soluble factors and cellular components, along with large quantities of inflammatory stroma, plays a critical role in PDAC occurrence and progression. Using genetically engineered mouse (GEM) models of PDAC, researchers have investigated the initial steps of pancreatic tumorigenesis in the context of the microenvironment and the role of KRAS in the microenvironment (7). Pancreatic stellate cells (PaSCs) have structures and functions similar to those of hepatic stellate cells and usually exist in the resting state. In the earliest stages of PanIN, mesenchymal cells are transformed into fibroblasts and pancreatic stellate cells upon injury induction. PaSC proliferation and activation are central to the development of pancreatic fibrosis. Activated PaSCs synthesize many extracellular matrix components (8), and infiltration of these cells can be influenced by KRAS gene activity.
The expression of KRAS-dependent factors, such as interleukin-6 (IL-6) and Sonic hedgehog (Shh), in the tumor microenvironment is modulated by KRAS (9). The Hedgehog signaling pathway plays a significant role in stromal desmoplasia in PDAC and accelerates the progression of oncogenic disease driven by KRAS (10). KRAS G12D upregulates Hedgehog signaling to mediate paracrine interactions in the microenvironment. Additionally, overexpression of the soluble ligand Shh stimulates PDAC cells and promotes the formation of desmoplastic stroma brimming with fibroblasts (11). In the iKRAS mouse model, when KRAS is inactivated, Shh expression in epithelial cells is decreased to regular levels (9). IL-6, an inflammatory cytokine that has been associated with PDAC development, plays a critical role in various biologic activities (12). The serum concentration of IL-6 is increased in PDAC, and its expression is enhanced by KRAS.
Metabolic reprogramming
The process of tumor cell metabolic reprogramming is one of the most typical mechanisms by which tumors to adapt to the microenvironment, maintain cell survival and meet the needs of macromolecule synthesis (13,14). In this sense, tumors are also considered a metabolic disease (15). Under hypoxia deficiency (16), PDAC metabolic processes exhibit significant changes (17). Moreover, the Warburg effect and altered mitochondrial metabolic activity are typical metabolic changes in pancreatic cancer (18,19). Even with sufficient oxygen, PDAC utilizes aerobic glycolysis to replace oxidative phosphorylation (OXPHOS) in normal tissue cells to provide energy, and this process is called the Warburg effect. In one aspect, KRAS adjusts to the metabolic changes in PDAC by increasing the expression of glycolytic enzymes, such as hexokinase 1 and 2, glucose transporters, phosphofructokinase and lactate dehydrogenase. On the other hand, the synthesis of proteins, nucleic acids and fatty acids needed for PDAC cell proliferation is supported by KRAS mainly through stimulation of glucose uptake and glucose mid products into the hexosamine and pentose phosphate pathways (20,21).
Another role of KRAS in promoting PDAC growth affects the glutamate metabolic pathway. Most cells require glutamate dehydrogenase to convert glutamine into α-ketoglutarate within the mitochondria to fuel the tricarboxylic acid cycle. However, in PDAC, glutamine-derived aspartate is transported to the cytoplasm and converted to oxaloacetate by aspartate aminotransferase, and oxaloacetate is subsequently converted to malic acid and finally becomes pyruvate, followed by an increase in the NADPH/NADP ratio to maintain the redox levels in PDAC cells. Pancreatic cancer cells are highly dependent on this metabolic pathway. KRAS mutations lead to increased aspartate aminotransferase expression and promote glutamine metabolism in PDAC, maintaining redox levels and promoting cancer cell growth (22).
Additionally, KRAS leads to metabolic changes that alter the generation of mitochondrial reactive oxygen species (ROS) (23). PDAC cells develop several mechanisms to resist the excessive ROS levels, which are detrimental to tumor cells. Therefore, the cancer cells can reduce cellular damage caused by ROS (24). Currently, KRAS mutation is considered to activate nuclear factor-erythroid 2-related factor 2 (Nrf2) to start the antioxidant mechanism (25), which activates a series of antioxidant genes. More than 100 genes have been reported to be regulated by Nrf2, including NADPH regulators, drug efflux pumps and growth factors (26). The inhibitor KEAP1 strictly controls Nrf2 levels by binding to Nrf2 and mediating Nrf2 ubiquitination; thus, under normal conditions, Nrf2 levels remain low (27). One study found that mutant KRAS signals mainly through the Mek-Erk-Jun pathway to promote Nrf2 nuclear localization and antioxidative gene expression (28). In pancreatic cancer, human PanIN and PDAC, Nrf2 is more active than in normal pancreatic ductal cells, and the ROS level remains low. By contrast, because of the effect of KEAP1, Nrf2 in normal pancreatic duct cells is maintained at a very low level. Unlike many other tumors, few somatic mutations lead to inactivation of NRF2 in pancreatic cancer (29). Therefore, in KRAS-mutant pancreatic cancer, Nrf2-mediated antioxidant activity is regulated mainly by KRAS mutation. By contrast, knockdown of KRAS or MAPK signaling blocks Nrf2 expression and increases intracellular ROS levels (28).
Early diagnosis
Compared with resectable PDAC, the prognosis of patients with unresectable PDAC is worse (30); thus, an early diagnosis is essential for these patients. Endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) can scan the lesion within a short range and has few complications. Because pathological diagnosis is still the gold standard for PDAC diagnosis, EUS-FNA has become the preferred method to diagnose pancreatic disease and has been increasingly used clinically (31). EUS-FNA can be used to detect gene mutations in tissues or cells obtained by aspiration (32). Combination of a KRAS-mutation assay with cytopathology is better than cytopathology alone in increasing the sensitivity, negative predictive value and accuracy in inconclusive or doubtful diagnoses based on cytopathology and thus is central to inconclusive or doubtful diagnoses from cytopathology. This KRAS-mutation detection approach cannot replace histology but complements histology (33), especially for patients with an indeterminate mass in the pancreas (34). However, EUS-FNA may lead to the release of cellular material from tumors to the bloodstream. The detection of mutant KRAS and the concentration of cfDNA in plasma are increased after the procedure (35). Thus, further studies are needed.
In addition to EUS-FNA, a new emerging detection method, cancer-specific DNA detection, was recently identified. This method, called a liquid biopsy, detects cancer-specific DNA in peripheral blood and noninvasively examines tumor characteristics. KRAS mutations can be detected both in plasma and serum DNA (36,37). Carbohydrate antigen 19-9 (CA19-9) is currently the most meaningful and widely used biomarker in PDAC, and has a sensitivity and specificity of 79–81% and 82–90% respectively. However, approximately 5% to 10% of the PDAC patients are Lewis negative individuals which are documented to have scarce or no CA19-9 secretion (38). Therefore, measurement of ctDNA for detection and quantitative monitoring of KRAS mutations may offer an additional diagnostic biomarker to CA19-9 (39). Baseline ctDNA KRAS detection rate was 93.7% (86.4% in patients with non-elevated CA19-9) with the application of an ultrasensitive ctDNA KRAS assay (40). However, the KRAS-mutation subtype in peripheral blood may be heterogeneous compared with that in the primary tumor (41). Additionally, KRAS mutations can be detected through genomic DNA in exosomes derived from the serum of PDAC patients. Exosomes are small vesicles (50–150 nm) of endocytic origin that are shed from viable cells into the circulation, and serum exosomes from patients with PDAC contain genomic DNA spanning all chromosomes (42). Exosomal DNA (exoDNA) can be detected even in the early stage of PDAC. Additionally, KRAS mutations in exoDNA were found in 7.4%, 66.7%, 80%, and 85% of age-matched controls and localized, locally advanced, and metastatic PDAC patients, respectively, which is higher than the detection levels in cfDNA (14.8%, 45.5%, 30.8%, and 57.9%, respectively) (43). However, mutant KRAS in the circulation has also been detected in healthy samples, possibly limiting its usefulness as a marker for early diagnosis.
Prognosis
There is still no consensus on whether KRAS mutations affect the prognosis of PDAC patients. Many studies have found that the presence of a KRAS mutation and a KRAS mutational subtype were both associated with a poor prognosis in PDAC patients (44,45). A single-nucleotide mutation induces a replacement of the GGT sequence (encoding glycine) by the GAT (aspartic acid-G12D-c35 G>A), GTT (valine-G12V-c35 G>T), CGT (arginine-G12R-C34 G>C), or GCT (alanine-G12A-c35 G>C) sequence. The KRAS G12 mutation accounts for 99% of all mutations. A point mutation can also happen on codon 13 (G13D) or 61 (Q61L or Q61H) but is less frequent (46,47). Whole-exome sequencing of PDAC revealed that codon Q61 alleles of KRAS are specifically associated with a better prognosis (48). Using next-generation sequencing, Qian et al. (49) analyzed DNA alterations in four main driver genes in 356 patients with resected PDAC. They found that patients who had KRAS-mutant tumors had a worse disease-free survival (DFS) and overall survival (OS) than those with KRAS wild-type tumors. Additionally, patients with KRAS G12D-mutant tumors had particularly different outcomes and the worst DFS. Ako et al. (36) proposed the contrasting idea that the prognosis of KRAS G12V-mutant tumors was poorer than other subtypes. In metastatic pancreatic cancer patients, KRAS G12V mutation was also correlated with poor OS based on subgroup analysis (50). Hamidi et al. (51) used a growth inhibition assay to determine the sensitivity to MEK inhibition in different KRAS mutational subtypes and copy number variations. They found that cell lines with KRAS G12V mutation and KRAS gain or loss were ~10 times more resistant than the other subtypes. Copy number variation may be an important biomarker for PDAC. Thus, multicenter investigations in a larger homogeneous cohort of PDAC patients are certainly needed in the future to reach a definitive conclusion.
Patients with KRAS wild-type tumors seem to benefit from chemotherapy. A multicenter, randomized phase IIb study found that patients with KRAS-mutant tumors experienced a significantly poorer prognosis than those with KRAS wild-type tumors when administered gemcitabine combined with the monoclonal antibody nimotuzumab (52). Another phase III trial that compared gemcitabine/erlotinib followed by capecitabine with capecitabine/erlotinib followed by gemcitabine in advanced PC also showed that KRAS wild-type patients have an improved OS (53). Early changes in the plasma DNA concentration of mutant KRAS is a sensitive index of the chemotherapy effect of the start stage in PDAC (54). An increase in plasma DNA in the sample collected from patients after chemotherapy was correlated with poorer PFS and OS than observed in patients with stable/reduced plasma DNA (55). The dynamic changes in ctDNA KRAS mutation load from serial measurements may be used as an assessment of therapeutic response, which was independent and complementary to the commonly used biomarker CA19-9, especially in Lewis negative individuals (40). And ctDNA level over time is a better predictor of survival than the dynamics of CA19-9 (56).
Therapeutics
Because the KRAS gene plays a vital role in PDAC development, it is an attractive therapeutic target. However, there is still no lone path to an effective treatment for KRAS-mutant PDAC (57). Nonetheless, researchers have attempted to find possible paths that can suppress the effects of KRAS in PDAC (58).
Many studies have tried to inhibit RAS directly. Burns et al. (59) reported the identification of a small molecule that can link to a unique pocket on the Ras:Son of Sevenless (SOS): Ras complex and increase the rate of SOS-catalyzed nucleotide exchange in vitro. This finding provides a new target for the discovery of potent Ras signaling inhibitors, but this complex has yet to be targeted by existing compounds (Figure 1).
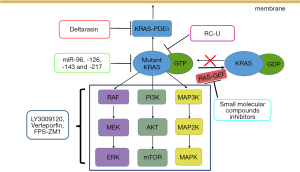
Compared with the protein inhibition approach, mRNA targeting via RNA interference (RNAi) has already been shown to be an effective alternative (60). Zorde Khvalevsky et al. (61) developed a local prolonged siRNA delivery system (Local Drug EluteR, LODER) that sheds siRNA against mutant KRAS (siG12D LODER) (Figure 1). They found that the in vitro growth of pancreatic cancer cells can be substantially inhibited by LODER-derived siG12D and that in vivo tumor growth can also be suppressed. Another phase 1/2a study showed that the combination of LODER-derived siG12D and chemotherapy demonstrates a potential treatment effect in patients with locally advanced pancreatic cancer (60). Additionally, many miRNAs have been shown to target KRAS in PDAC, including let-7a, miR-96, miR-126, miR-143 and miR-217 (62,63). For example, miR-126 can directly target KRAS at a ‘seedless’ binding site within its 3´ UTR. Replacing these miRNAs in KRAS-mutant PDAC patients may represent a new approach to preventing tumor progression and metastasis (62).
RAS must be positioned at the cell membrane to maintain its biological activity, and this association is induced by farnesyl transferase (FTase) (64). FTase attaches a 15-carbon farnesyl isoprenoid to the cysteine in the CAAX-motif, which is the first step in the CAXX modifications. Some farnesyltransferase inhibitors (FTIs), such as tipifarnib, have been tested clinically but do not show clinical benefit (65,66), likely because the RAS gene encodes four different proteins—HRAS, NRAS, KRAS4a, and KRAS4b—and because HRAS and NRAS do not rely on farnesylation. Another approach to interfere with RAS membrane association is blocking the RAS pathway to the plasma membrane transporters. Phosphodiesterase 6 delta (PDE δ) has a pocket that can combine with RAS protein that has been modified by FTase. PDE δ can promote RAS protein distribution in the correct position and is involved in signal transduction (67). Deltarasin is a small-molecule inhibitor that inhibits the KRAS-PDEδ interaction. Through this process, oncogenic RAS signaling and the oncogenic KRAS-dependent proliferation of human PDAC cells are suppressed (Figure 1) (68). This brings a new approach to suppressing oncogenic RAS signaling.
Numerous inhibitors that target RAS downstream effector signaling (Figure 1), such as RAF/MEK/ERK and PI3K/AKT/mTOR signaling pathways, are already being tested in clinical trials. Many inhibitor combinations for PDAC treatment have demonstrated promising results (69). LY3009120 is a pan-RAF inhibitor with activity against three RAF isoforms and avoids the induction of paradoxical downstream signaling activation (70). Verteporfin, a YAP inhibitor (71), can block the activation of a parallel AKT signaling pathway after LY3009120 treatment, significantly enhancing the antitumor efficacy of LY3009120 (72). Many inhibitor-based combinations must be informed by the activation state of each putative driver in a given treatment context (73) and will be needed for efficacy across different KRAS-mutant PDAC populations (74,75). Some inhibitors can block the ERK signaling pathway indirectly. For example, inhibition of receptor for advanced glycation end-products (RAGE) using the pharmacological antagonist FPS-ZM1 restrained ERK activity downstream of KRAS in PDAC cell lines (76).
Chimeric antigen receptor T cells (CAR-T) have shown huge success against CD19-ex-pressing B cell leukaemia (77). As CAR T cell therapy spread, so does the search for new biomarker targets for PDAC (78). However, there are reports on the CAR-T cells treatment which target an upstream component of the RAS signaling pathway (i.e., EGFR/HER2) in pancreatic cancer. Raj et al. (79) used switchable CAR-T cells to target the antigen HER2. They found that a switchable CAR-T system is effective against aggressive and disseminated tumours derived from patients with advanced PDAC while affording the potential safety of a control switch.
In addition, many alternative strategies exist for targeted KRAS therapy in PDAC. Ma et al. (80) generated an engineered E3 ubiquitin ligase (RC-U) to target the KRAS oncoprotein for ubiquitination and degradation, resulting in a reduction in PDAC cell proliferation both in vitro and in vivo (Figure 1). Exosomes that carry short interfering RNA or short hairpin RNA specific to KRAS G12D, generated by Kamerkar, have been confirmed to significantly suppress cancer development in multiple mouse models (81).
Summary
KRAS can be used to predict the prognosis and assist in the early diagnosis of PDAC. Unfortunately, despite the many paths available to suppress the effects of KRAS, none of the treatments targeting these paths have been significantly successful in the clinic. More studies are required to further elucidate the effects of KRAS and identify therapeutic targets in PDAC.
Acknowledgments
Funding: This work was supported by The National Science Fund for Distinguished Young Scholars [81625016].
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.10.19). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Lin HJ, Lin J. Seed-in-soil: Pancreatic cancer influenced by tumor microenvironment. Cancers (Basel) 2017;9: [PubMed]
- Cicenas J, Kvederaviciute K, Meskinyte I, et al. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 Mutations in Pancreatic Cancer. Cancers (Basel) 2017;9: [PubMed]
- Giri B, Sethi V, Dudeja V, et al. Genetics of pancreatic cyst-cancer progression: Standing on the shoulders of giants. Curr Opin Gastroenterol 2017;33:404-10. [Crossref] [PubMed]
- Kanda M, Matthaei H, Wu J, et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 2012;142:730-733.e9. [Crossref] [PubMed]
- Cicenas J, Tamosaitis L, Kvederaviciute K, et al. KRAS, NRAS and BRAF mutations in colorectal cancer and melanoma. Med Oncol 2017;34:26. [Crossref] [PubMed]
- Guerra C, Barbacid M. Genetically engineered mouse models of pancreatic adenocarcinoma. Mol Oncol 2013;7:232-47. [Crossref] [PubMed]
- Won JH, Zhang Y, Ji B, et al. Phenotypic changes in mouse pancreatic stellate cell ca2+ signaling events following activation in culture and in a disease model of pancreatitis. Mol Biol Cell 2011;22:421-36. [Crossref] [PubMed]
- Collins MA, Bednar F, Zhang Y, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest 2012;122:639-53. [Crossref] [PubMed]
- Lee JJ, Perera RM, Wang H, et al. Stromal response to hedgehog signaling restrains pancreatic cancer progression. Proc Natl Acad Sci U S A 2014;111:E3091-100. [Crossref] [PubMed]
- Liu X, Pitarresi JR, Cuitino MC, et al. Genetic ablation of smoothened in pancreatic fibroblasts increases acinar-ductal metaplasia. Genes Dev 2016;30:1943-55. [Crossref] [PubMed]
- Holmer R, Goumas FA, Waetzig GH, et al. Interleukin-6: A villain in the drama of pancreatic cancer development and progression. Hepatobiliary Pancreat Dis Int 2014;13:371-80. [Crossref] [PubMed]
- Kerr EM, Gaude E, Turrell FK, et al. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature 2016;531:110-3. [Crossref] [PubMed]
- Robey RB, Weisz J, Kuemmerle NB, et al. Metabolic reprogramming and dysregulated metabolism: Cause, consequence and/or enabler of environmental carcinogenesis? Carcinogenesis 2015;36:S203-231. [Crossref] [PubMed]
- Oronsky BT, Oronsky N, Fanger GR, et al. Follow the ATP: Tumor energy production: A perspective. Anticancer Agents Med Chem 2014;14:1187-98. [Crossref] [PubMed]
- Makohon-Moore A, Iacobuzio-Donahue CA. Pancreatic cancer biology and genetics from an evolutionary perspective. Nat Rev Cancer 2016;16:553-65. [Crossref] [PubMed]
- Seton-Rogers S. Pancreatic cancer: A matter of timing. Nat Rev Cancer 2015;15:256-7. [Crossref] [PubMed]
- Olivares O, Vasseur S. Metabolic rewiring of pancreatic ductal adenocarcinoma: New routes to follow within the maze. Int J Cancer 2016;138:787-96. [Crossref] [PubMed]
- Gasparre G, Porcelli AM, Lenaz G, et al. Relevance of mitochondrial genetics and metabolism in cancer development. Cold Spring Harb Perspect Biol 2013;5: [Crossref] [PubMed]
- Ying H, Kimmelman AC, Lyssiotis CA, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 2012;149:656-70. [Crossref] [PubMed]
- Sousa CM, Kimmelman AC. The complex landscape of pancreatic cancer metabolism. Carcinogenesis 2014;35:1441-50. [Crossref] [PubMed]
- Son J, Lyssiotis CA, Ying H, et al. Glutamine supports pancreatic cancer growth through a Kras-regulated metabolic pathway. Nature 2013;496:101-5. [Crossref] [PubMed]
- Storz P. KRAS, ROS and the initiation of pancreatic cancer. Small GTPases 2017;8:38-42. [Crossref] [PubMed]
- Rosenfeldt MT, O'Prey J, Morton JP, et al. P53 status determines the role of autophagy in pancreatic tumour development. Nature 2013;504:296-300. [Crossref] [PubMed]
- DeNicola GM, Karreth FA, Humpton TJ, et al. Oncogene-induced nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011;475:106-9. [Crossref] [PubMed]
- Hayes JD, McMahon M. Nrf2 and keap1 mutations: Permanent activation of an adaptive response in cancer. Trends Biochem Sci 2009;34:176-88. [Crossref] [PubMed]
- Perera RM, Bardeesy N. Cancer: When antioxidants are bad. Nature 2011;475:43-4. [Crossref] [PubMed]
- Todoric J, Antonucci L, Di Caro G, et al. Stress-activated nrf2-mdm2 cascade controls neoplastic progression in pancreas. Cancer Cell 2017;32:824-839.e8. [Crossref] [PubMed]
- Chio II, Jafarnejad SM, Ponz-Sarvise M, et al. Nrf2 promotes tumor maintenance by modulating MRNA translation in pancreatic cancer. Cell 2016;166:963-76. [Crossref] [PubMed]
- Vincent A, Herman J, Schulick R, et al. Pancreatic cancer. Lancet 2011;378:607-20. [Crossref] [PubMed]
- Hamada T, Yasunaga H, Nakai Y, et al. Severe bleeding and perforation are rare complications of endoscopic ultrasound-guided fine needle aspiration for pancreatic masses: An analysis of 3,090 patients from 212 hospitals. Gut Liver 2014;8:215-8. [Crossref] [PubMed]
- Bournet B, Buscail C, Muscari F, et al. Targeting KRAS for diagnosis, prognosis, and treatment of pancreatic cancer: Hopes and realities. Eur J Cancer 2016;54:75-83. [Crossref] [PubMed]
- Bournet B, Selves J, Grand D, et al. Endoscopic ultrasound-guided fine-needle aspiration biopsy coupled with a KRAS mutation assay using allelic discrimination improves the diagnosis of pancreatic cancer. J Clin Gastroenterol 2015;49:50-6. [Crossref] [PubMed]
- Wang X, Gao J, Ren Y, et al. Detection of KRAS gene mutations in endoscopic ultrasound-guided fine-needle aspiration biopsy for improving pancreatic cancer diagnosis. Am J Gastroenterol 2011;106:2104-11. [Crossref] [PubMed]
- Levy MJ, Kipp BR, Milosevic D, et al. Analysis of cell-free DNA to assess risk of tumoremia following endoscopic ultrasound fine-needle aspiration of pancreatic adenocarcinomas. Clin Gastroenterol Hepatol 2018;16:1632-1640.e1. [Crossref] [PubMed]
- Ako S, Nouso K, Kinugasa H, et al. Utility of serum DNA as a marker for KRAS mutations in pancreatic cancer tissue. Pancreatology 2017;17:285-90. [Crossref] [PubMed]
- Yang C, Luo G, Cheng H, et al. Potential biomarkers to evaluate therapeutic response in advanced pancreatic cancer. Transl Cancer Res 2018;7:208-18. [Crossref]
- Luo G, Liu C, Guo M, et al. Potential biomarkers in Lewis negative patients with pancreatic cancer. Ann Surg 2017;265:800-5. [Crossref] [PubMed]
- Sefrioui D, Blanchard F, Toure E, et al. Diagnostic value of ca19.9, circulating tumour DNA and circulating tumour cells in patients with solid pancreatic tumours. Br J Cancer 2017;117:1017-25. [Crossref] [PubMed]
- Chen I, Raymond VM, Geis JA, et al. Ultrasensitive plasma ctDNA KRAS assay for detection, prognosis, and assessment of therapeutic response in patients with unresectable pancreatic ductal adenocarcinoma. Oncotarget 2017;8:97769-86. [PubMed]
- Kulemann B, Rosch S, Seifert S, et al. Pancreatic cancer: Circulating tumor cells and primary tumors show heterogeneous KRAS mutations. Sci Rep 2017;7:4510. [Crossref] [PubMed]
- Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem 2014;289:3869-75. [Crossref] [PubMed]
- Allenson K, Castillo J, San Lucas FA, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol 2017;28:741-7. [PubMed]
- Diep CH, Munoz RM, Choudhary A, et al. Synergistic effect between erlotinib and MEK inhibitors in KRAS wild-type human pancreatic cancer cells. Clin Cancer Res 2011;17:2744-56. [Crossref] [PubMed]
- Bournet B, Muscari F, Buscail C, et al. KRAS G12D mutation subtype is a prognostic factor for advanced pancreatic adenocarcinoma. Clin Transl Gastroenterol 2016;7:e157 [Crossref] [PubMed]
- di Magliano MP, Logsdon CD. Roles for KRAS in pancreatic tumor development and progression. Gastroenterology 2013;144:1220-9. [Crossref] [PubMed]
- Forbes SA, Bindal N, Bamford S, et al. Cosmic: Mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res 2011;39:D945-50. [Crossref] [PubMed]
- Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun 2015;6:6744. [Crossref] [PubMed]
- Qian ZR, Rubinson DA, Nowak JA, et al. Association of alterations in main driver genes with outcomes of patients with resected pancreatic ductal adenocarcinoma. JAMA Oncol 2018;4:e173420 [Crossref] [PubMed]
- Cheng H, Liu C, Jiang J, et al. Analysis of ctDNA to predict prognosis and monitor treatment responses in metastatic pancreatic cancer patients. Int J Cancer 2017;140:2344-50. [Crossref] [PubMed]
- Hamidi H, Lu M, Chau K, et al. KRAS mutational subtype and copy number predict in vitro response of human pancreatic cancer cell lines to MEK inhibition. Br J Cancer 2014;111:1788-801. [Crossref] [PubMed]
- Schultheis B, Reuter D, Ebert MP, et al. Gemcitabine combined with the monoclonal antibody nimotuzumab is an active first-line regimen in KRAS wildtype patients with locally advanced or metastatic pancreatic cancer: A multicenter, randomized phase IIb study. Ann Oncol 2017;28:2429-35. [Crossref] [PubMed]
- Boeck S, Jung A, Laubender RP, et al. KRAS mutation status is not predictive for objective response to anti-EGFR treatment with erlotinib in patients with advanced pancreatic cancer. J Gastroenterol 2013;48:544-8. [Crossref] [PubMed]
- Earl J, Garcia-Nieto S, Martinez-Avila JC, et al. Circulating tumor cells (CTC) and kras mutant circulating free DNA (cfDNA) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer 2015;15:797. [Crossref] [PubMed]
- Del Re M, Vivaldi C, Rofi E, et al. Early changes in plasma DNA levels of mutant KRAS as a sensitive marker of response to chemotherapy in pancreatic cancer. Sci Rep 2017;7:7931. [Crossref] [PubMed]
- Perets R, Greenberg O, Shentzer T, et al. Mutant KRAS circulating tumor DNA is an accurate tool for pancreatic cancer monitoring. Oncologist 2018;23:566-72. [Crossref] [PubMed]
- Zeitouni D, Pylayeva-Gupta Y, Der CJ, et al. KRAS mutant pancreatic cancer: No lone path to an effective treatment. Cancers (Basel) 2016;8: [Crossref] [PubMed]
- Berger NA. Actionable intelligence provided by pancreatic cancer genomic landscape: Are targets for curative therapy on the map? Transl Cancer Res 2016;5:S243-S247. [Crossref] [PubMed]
- Burns MC, Sun Q, Daniels RN, et al. Approach for targeting RAS with small molecules that activate SOS-mediated nucleotide exchange. Proc Natl Acad Sci U S A 2014;111:3401-6. [Crossref] [PubMed]
- Golan T, Khvalevsky EZ, Hubert A, et al. RNAI therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget 2015;6:24560-70. [Crossref] [PubMed]
- Zorde Khvalevsky E, Gabai R, Rachmut IH, et al. Mutant KRAS is a druggable target for pancreatic cancer. Proc Natl Acad Sci U S A 2013;110:20723-8. [Crossref] [PubMed]
- Frampton AE, Krell J, Jacob J, et al. Loss of mir-126 is crucial to pancreatic cancer progression. Expert Rev Anticancer Ther 2012;12:881-4. [Crossref] [PubMed]
- Yu S, Lu Z, Liu C, et al. Mirna-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res 2010;70:6015-25. [Crossref] [PubMed]
- Ahearn IM, Haigis K, Bar-Sagi D, et al. Regulating the regulator: Post-translational modification of RAS. Nat Rev Mol Cell Biol 2011;13:39-51. [Crossref] [PubMed]
- Macdonald JS, Mccoy S, Whitehead RP, et al. A phase II study of farnesyl transferase inhibitor r115777 in pancreatic cancer: A southwest oncology group (SWOG 9924) study. Investigational New Drugs 2005;23:485-7. [Crossref] [PubMed]
- Heestand GM, Kurzrock R. Molecular landscape of pancreatic cancer: Implications for current clinical trials. Oncotarget 2015;6:4553-61. [Crossref] [PubMed]
- Chandra A, Grecco HE, Pisupati V, et al. The GDI-like solubilizing factor PDE ´ sustains the spatial organization and signalling of RAS family proteins. Nature Cell Biology 2011;14:148-58. [Crossref] [PubMed]
- Zimmermann G, Papke B, Ismail S, et al. Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature 2013;497:638-42. [Crossref] [PubMed]
- Ischenko I, Petrenko O, Hayman MJ A. MEK/pi3k/HDAC inhibitor combination therapy for KRAS mutant pancreatic cancer cells. Oncotarget 2015;6:15814-27. [Crossref] [PubMed]
- Peng SB, Henry JR, Kaufman MD, et al. Inhibition of RAF isoforms and active dimers by ly3009120 leads to anti-tumor activities in RAS or BRAF mutant cancers. Cancer Cell 2015;28:384-98. [Crossref] [PubMed]
- Iacono P, Tedeschi M, Boccassini B, et al. Chronic central serous chorioretinopathy: Early and late morphological and functional changes after verteporfin photodynamic therapy. Retina 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Zhao X, Wang X, Fang L, et al. A combinatorial strategy using YAP and pan-RAF inhibitors for treating KRAS-mutant pancreatic cancer. Cancer Letters 2017;402:61-70. [Crossref] [PubMed]
- Pettazzoni P, Viale A, Shah P, et al. Genetic events that limit the efficacy of MEK and RTK inhibitor therapies in a mouse model of KRAS-driven pancreatic cancer. Cancer Res 2015;75:1091-101. [Crossref] [PubMed]
- Hayes TK, Neel NF, Hu C, et al. Long-term ERK inhibition in KRAS-mutant pancreatic cancer is associated with MYC degradation and senescence-like growth suppression. Cancer Cell 2016;29:75-89. [Crossref] [PubMed]
- Basseville A, Bates S, Fojo T. Pancreatic cancer: Targeting KRAS and the vitamin d receptor via microtubules. Nat Rev Clin Oncol 2015;12:442-4. [Crossref] [PubMed]
- Azizan N, Suter MA, Liu Y, et al. Rage maintains high levels of NFκB and oncogenic Kras activity in pancreatic cancer. Biochem Biophys Res Commun 2017;493:592-7. [Crossref] [PubMed]
- Kalos M, Levine BL, Porter DL, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med 2011;3:95ra73 [Crossref] [PubMed]
- Townsend MH, Shrestha G, Robison RA, et al. The expansion of targetable biomarkers for car t cell therapy. J Exp Clin Cancer Res 2018;37:163. [Crossref] [PubMed]
- Raj D, Yang MH, Rodgers D, et al. Switchable car-t cells mediate remission in metastatic pancreatic ductal adenocarcinoma. Gut 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Ma Y, Gu Y, Zhang Q, et al. Targeted degradation of KRAS by an engineered ubiquitin ligase suppresses pancreatic cancer cell growth in vitro and in vivo. Mol Cancer Ther 2013;12:286-94. [Crossref] [PubMed]
- Kamerkar S, LeBleu VS, Sugimoto H, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017;546:498-503. [PubMed]


