
LncRNA NKILA suppresses colon cancer cell proliferation and migration by inactivating PI3K/Akt pathway
Introduction
As the most common type of gastrointestinal malignancy, colon cancer is considered to be the fourth major cause of cancer-related deaths worldwide (1). At present, colon cancer affects 12 million new cases and causes more than half million deaths every year (2). Incidence rate of colon cancer increases with age, and is higher in men than in women (3). With progresses in treatment of colon cancer, 5-year survival rate of colon cancer patients without distant metastasis now has reached 90% in some developed countries, however, once distant metastasis occurs, the 5-year survival rate will be as low as 10% (4). Therefore, early diagnosis and treatment is the key for survival of colon cancer patients.
Long non-coding RNA, also known as lncRNA, is a group of non-coding RNA containing more than 200 nucleotides (5). Numerous studies have shown that lncRNAs play pivotal roles in nearly all aspects of normal biological processes in the human body (6). Besides that, many lncRNAs are also involved in pathogenesis of a variety of human diseases including heart diseases, liver diseases and different types of human cancers (7-9). As a lncRNA that can be induced by NF-ĸB pathway-related cytokines (10), NKILA plays a role as tumor suppressor gene in various types of human cancers including non-small cell lung cancer (11) and tongue squamous cell carcinoma (12). However, functionality of this lncRNA in colon cancer still hasn’t been reported.
In this study, expression of NKILA in tumor tissues of patients with different stages of colon cancer was detected. Effects of NKILA knockdown and overexpression on migration and invasion of cells of different colorectal adenocarcinoma cell lines were studied, and the interaction between NKILA and PI3K/Akt pathway was also explored. The report is as follow:
Methods
Patients
A total of 100 patients with colon cancer were selected in The First Affiliated Hospital of Kunming Medical University from January 2013 to January 2016. All patients were diagnosed by pathological tests and imaging examinations. Those patients were divided into different stages according to the standard established by American Joint Committee on Cancer: stage 0, primary tumor without lymph node metastasis and distant metastasis; stage 1, submucosal layer or muscle layer was invaded, but no lymph node metastasis or distant metastasis was observed; stage 2, large intestine without peritoneal coverage was invaded, but no lymph node metastasis and distant metastasis were observed; stage 3, other organs were invaded, lymph node metastasis was observed, but no distant metastasis was found; stage 4, other organs were invaded, lymph node metastasis and distant metastasis were found. Each stage contained 20 patients. Patients in stage 0 included 12 males and 8 females, and age ranged from 25 to 61 years, with a mean age of 44.8±12.1 years; patients in stage 1 included 11 males and 9 females, and the age ranged from 31 to 72 years, with a mean age of 49.1±9.1 years; patients in stage 2 included 9 males and 11 females, and age ranged from 19 to 65 years, with a mean age of 42.2±17.1 years; patients in stage 3 included 8 males and 12 females, and age ranged from 20 to 71 years, with a mean age of 45.6±11.3 years; patients in stage 3 included 10 males and 10 females, and age ranged from 33 to 72 years, with a mean age of 40.5±16.1 years. There were no significant differences in age, gender and other basic information among patients at different stages. Surgical resection was performed for all patients, and tumor tissues were collected during operation. All participants signed informed consent. This study was approved by ethics committee of The First Affiliated Hospital of Kunming Medical University.
Cell lines, cell culture and transfection
Colorectal adenocarcinoma cell lines HT-29, HCT-8 and C2BBe1, and normal cell line CCD-18 Co were purchased from ATCC. All cell lines were cultured according to the manufacture’s instructions. Cells were collected during logarithmic growth phase for all subsequent experiments. Transfection through lentivirus was performed according to the method described by Huang et al. (12).
Real-time quantitative PCR
Total RNA was extracted from tumor tissues and cells of different cell lines using Trizol reagent (Invitrogen, USA). NanoDrop™ 2000 Spectrophotometers (Thermo Fisher Scientific, USA) was used to check the quality of all RNA samples, and only the ones with a ratio of A260/A280 between 1.8 and 2.0 were used in reverse transcription. SYBR® Green Real-Time PCR Master Mixes (Thermo Fisher Scientific, USA) was used to prepare PCR reaction systems. Following primers were used in PCR reactions: 5'-TGGATTGTTGGGTATATTTTGGA-3' (forward) and 5'-TGTATGAAGAGGATGCTGAAGGC-3' (reverse) for NKILA; 5'-GACCTCTATGCCAACACAGT-3' (forward) and 5'-AGTACTTGCGCTCAGGAGGA-3' (reverse) for β-actin. PCR reaction conditions were: 95 °C for 1 min, followed by 40 cycles of 95 °C for 20 s and 60 °C for 50 s. Ct values were processed using 2−ΔΔCT method.
Transwell cell migration and invasion assay
Transwell cell migration and invasion assay (BD Biosciences, USA) was performed to investigate the migration and invasion ability cells of each cell line. For migration assay, about 4×104 cells were inoculated into the upper chamber, the lower chamber was filled with RPMI-1640 medium (Thermo Fisher Scientific, USA) supplemented with 20% FCS (Sigma-Aldrich, USA). After incubation for 24 h, membranes were stained with 0.5% crystal violet for 20 minutes. Results were observed under a light microscope (Olympus, Japan). Upper chamber was pre-coated with Matrigel (356234, Millipore, USA) before invasion assay, and all other steps were essentially the same with migration assay.
Western-blot
After protein extraction using conventional method, 12% SDS-PAGE gel electrophoresis was performed to separate different proteins. After transmembrane to PVDF, membranes were blocked with 5% skimmed milk, followed by incubation with primary antibodies including rabbit anti-pAkt (Thr308) (1:2000, ab38449, Abcam), anti-pAkt (Ser473) (1:2000, ab81283, Abcam), anti-Akt (1:2000, ab126811, Abcam), anti-PI 3 Kinase p85 alpha (pPI3K) (phospho Y607) (1:1000, ab182651, Abcam), anti-PI 3 Kinase p85 alpha (PI3K) and anti-GAPDH (1:1000, ab9845, Abcam) overnight at 4 °C. After washing, membranes were further incubated with anti-rabbit IgG-HRP secondary antibody (1:1000, MBS435036, MyBioSource) at room temperature for 2h. ECL method (Sigma-Aldrich, USA) was used to develop signals.
Mouse xenograft model
A total of 30 BALB/c nude mice (20±2 g) were provided by Guangdong Medical Laboratory Animal Center (Guangdong, China). Mice were randomly divided into 3 groups with 10 mice in each group. Normal HT-29 cells, HT-29 cells with NKILA overexpression and HT-29 cells with NKILA siRNA interference were resuspended in 100 µL of matrigel/PBS mixture and were subcutaneously injected into mice in different groups. Mice were sacrificed 6 weeks later. Tumors were collected and photographed. Animal experiments were performed following Guide for the Care and Use of Laboratory Animals-Chinese Version [1996]. Animal ethics committee of First Affiliated Hospital of Kunming Medical University approved this study.
Statistical analysis
PSS19.0 statistical software (SPSS Inc., USA) was used. All experiments were performed in triplicate manner. Measurement data were recorded by (
Results
Expression of lncRNA NKILA in patients with different stages of colon cancer
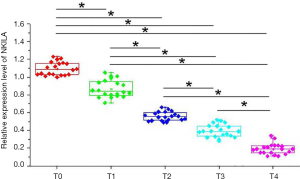
Expression of NKILA in tumor tissues of patients with different stages of colon cancer was detected by qRT-PCR. As shown in Figure 1, expression level of NKILA was significantly reduced with the progression of colon cancer (P<0.05). These data suggest that NKILA is involved in the development of colon cancer.
Expression of lncRNA NKILA in different cell lines with different treatment
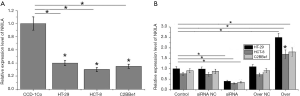
Colorectal adenocarcinoma accounts for about 95% of colon cancer. Therefore, three colorectal adenocarcinoma cell lines including HT-29, HCT-8 and C2BBe1 were used in this study. As shown in Figure 2A, compared with normal cell line CCD-18Co, expression of NKILA was significantly reduced in cells of all three colorectal adenocarcinoma cell lines (P<0.05). NKILA knockdown and overexpression cell lines were established through lentivirus infection. As shown in Figure 2B, expression level of NKILA was significantly reduced in cells of NKILA knockdown cell lines (P<0.05), and significantly increased in cells of NKILA overexpression cell lines (P<0.05), indicating the successfully established NKILA knockdown and overexpression cell lines.
Effects of NKILA knockdown and overexpression on migration and invasion of different colorectal adenocarcinoma cell lines
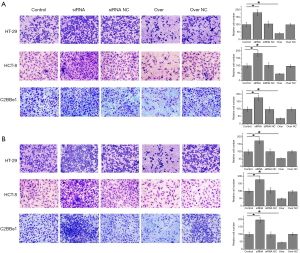
Transwell migration and invasion assay was performed to investigate the effects of NKILA knockdown and overexpression on migration and invasion abilities of cells of different colorectal adenocarcinoma cell lines. As shown in Figure 3A, NKILA knockdown through siRNA interference significantly increased the migration ability of cells of all three colorectal adenocarcinoma cell lines, while NKILA overexpression significantly decreased the migration ability of cells of all three colorectal adenocarcinoma cell lines (P<0.05). Similarly, as shown in Figure 3B, NKILA knockdown significantly increased the invasion ability of cells of all three colorectal adenocarcinoma cell lines, while NKILA overexpression significantly decreased the invasion ability (P<0.05). Those data suggest that expression level of NKILA is reversely correlated with the migration and invasion abilities of colorectal adenocarcinoma cells.
Effects of NKILA knockdown and overexpression on PI3K/Akt pathway
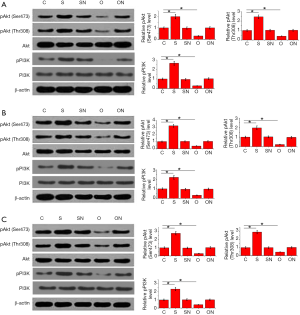
PI3K/Akt pathway plays essential roles in development of various cancers including colon cancer. Therefore, interaction between NKILA and PI3K/Akt pathway was explored in this study. As shown in Figure 4, NKILA knockdown and overexpression showed no significant effects on levels of total Akt and PI3K, while NKILA knockdown increased the levels of pAkt (Thr308), pAkt (Ser473) and pPI3K in cells of HT-29 (Figure 4A), HCT-8 (Figure 4B) and C2BBe1 (Figure 4C) three colorectal adenocarcinoma cell lines (P<0.05). In contrast, NKILA overexpression decreased the levels of pAkt (Thr308), pAkt (Ser473) and pPI3K in cells of these 3 colorectal adenocarcinoma cell lines (P<0.05). Those data suggest that NKILA expression can inhibit the activation of PI3K/Akt pathway.
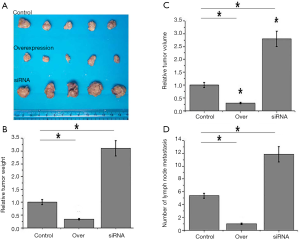
Effects of lncRNA NKILA overexpression and knockdown on the growth of tumors derived from colorectal adenocarcinoma cell line HT-29
As shown in Figure 5, compared with the tumors derived from control HT-29 cells, volume and weight of tumors derived from NKILA overexpressing HT-29 cells were significantly reduced (P<0.05). In contrast, the volume and weight of tumors derived from NKILA knockdown HT-29 cells were significantly increased (P<0.05). Those data suggest that NKILA can inhibit the growth of colon cancer.
Discussion
Colon cancer is usually considered as a group of heterogeneous diseases, and more than 30% cases of this disease are caused by genetic factors (13). Deletion or mutation of tumor suppressor genes and oncogenes in epithelial cells of gastrointestinal tract are major causes of colorectal adenocarcinoma, which is the dominant type of colon cancer (14). Recent advances in the pathogenesis of colon cancer also revealed the involvement of different lncRNAs. Liang et al. have shown that lncRNA H19 plays a role as oncogene to promote the epithelial to mesenchymal transition during the development of colon cancer by sequestering miRNAs (15). In another study, lncRNA HOTAIR was found to be upregulated during the progression of colorectal cancer, and the increased expression level of this lncRNA was found to be closely correlated with the poor prognosis (16). As a tumor suppressor gene, lncRNA NKILA was usually downregulated during the progression of different human malignant tumors such as breast cancer (17). Based on our knowledge, no studies on the expression pattern of NKILA in the development of colon cancer have been reported. In this study, expression level of NKILA was found to be significantly reduced with the progression of colon cancer, indicating that NKILA may also play a role as tumor suppressor gene in colon cancer.
Functionality of NKILA has been well studied in several human cancers. In the study of non-small cell lung cancer, NKILA was proved to inhibit the invasion and migration of tumor cells through interactions with NF-κB/Snail pathway (11). In another study, NKILA was reported to play as role as tumor suppressor in the development of tongue squamous cell carcinoma by inhibiting epithelial-mesenchymal transition (12). In the study of malignant melanoma, Bian et al. reported that NKILA could inhibit the development of tumor by regulating NF-ĸB signaling transduction pathway, indicating that NKILA may serve as a potential therapeutic target for the treatment of melanoma (18). In our study, increased invasion and migration abilities of different colorectal adenocarcinoma cell lines, including HT-29, HCT-8 and C2BBe1, were observed after NKILA knockdown. In addition, NKILA knockdown was also promoted the growth of tumor in mice derived from HT-29 cells. But NKILA overexpression played an opposite role. Those data suggest that NKILA can inhibit the growth of colon cancer by inhibiting the migration and invasion of tumor cells.
PI3K/Akt pathway was found to play pivotal roles in oncogenic transformation of various human cancers (19,20). Besides that, the activation of PI3K/Akt pathway was also found to be responsible for failures in treatment of cancers (21). Activation of PI3K/Akt was also observed in colon cancer (22). LncRNA NKILA may interact with NF-ĸB pathway to perform its functions (23). However, the interaction between NKILA and PI3K/AKT pathway still hasn’t been reported. In this study, increased levels of both pAkt (Thr308), pAkt (Ser473) and pPI3K in all three colorectal adenocarcinoma cell lines were observed after NKILA knockdown, while levels of both pAkt (Thr308), pAkt (Ser473) and Ppi3k decreased significantly after NKILA overexpression. Those data suggest that NKILA expression can inhibit the activation of PI3K/Akt pathway, which in turn inhibits the progression of tumor.
In conclusion, expression level of NKILA in patients was reduced with the development of colon cancer. NKILA can inhibit the migration and invasion of different colorectal adenocarcinoma cells by inhibiting the activation of PI3K/Akt pathway.
Acknowledgments
Funding: This work was supported by the Basic Application Fund of Yunnan Province [grant number 2015FB039].
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.11.05). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of The First Affiliated Hospital of Kunming Medical University. Signed written informed consents were obtained from the participants and/or guardians.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel R, DeSantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin 2014;64:104-17. [Crossref] [PubMed]
- Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet 2014;383:1490-502. [Crossref] [PubMed]
- Song M, Chan AT, Fuchs CS, et al. Dietary intake of fish, ω‐3 and ω‐6 fatty acids and risk of colorectal cancer: A prospective study in US men and women. Int J Cancer 2014;135:2413-23. [Crossref] [PubMed]
- Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin 2012;62:220-41. [Crossref] [PubMed]
- Perkel JM. Visiting" noncodarnia Biotechniques 2013;54:301-303-4. [Crossref] [PubMed]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011;12:861-74. [Crossref] [PubMed]
- Wang K, Liu F, Zhou LY, et al. The Long Noncoding RNA CHRF Regulates Cardiac Hypertrophy by Targeting miR-489Novelty and Significance. Circ Res 2014;114:1377-88. [Crossref] [PubMed]
- Takahashi K, Yan I, Haga H, et al. Long noncoding RNA in liver diseases. Hepatology 2014;60:744-53. [Crossref] [PubMed]
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 2011;1:391-407. [Crossref] [PubMed]
- Weidle UH, Birzele F, Kollmorgen G, et al. Long Non-coding RNAs and their Role in Metastasis. Cancer Genomics Proteomics 2017;14:143-60. [Crossref] [PubMed]
- Lu Z, Li Y, Wang J, et al. Long non-coding RNA NKILA inhibits migration and invasion of non-small cell lung cancer via NF-κB/Snail pathway. J Exp Clin Cancer Res 2017;36:54. [Crossref] [PubMed]
- Huang W, Cui X, Chen J, et al. Long non-coding RNA NKILA inhibits migration and invasion of tongue squamous cell carcinoma cells via suppressing epithelial-mesenchymal transition. Oncotarget 2016;7:62520-32. [PubMed]
- Dunlop MG. Colorectal cancer genetics. Semin Cancer Biol 1992;3:131-40. [PubMed]
- De Rosa M, Pace U, Rega D, et al. Genetics, diagnosis and management of colorectal cancer. Oncol Rep 2015;34:1087-96. [Crossref] [PubMed]
- Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 2015;6:22513-25. [Crossref] [PubMed]
- Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011;71:6320-6. [Crossref] [PubMed]
- Liu B, Sun L, Liu Q, et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015;27:370-81. [Crossref] [PubMed]
- Bian D, Gao C, Bao K, et al. The long non-coding RNA NKILA inhibits the invasion-metastasis cascade of malignant melanoma via the regulation of NF-ĸB. Am J Cancer Res 2017;7:28-40. [PubMed]
- Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis 2004;9:667-76. [Crossref] [PubMed]
- Chang F, Lee JT, Navolanic PM, et al. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia 2003;17:590-603. [Crossref] [PubMed]
- Lee S, Choi EJ, Jin C, et al. Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA amplification contributes to cisplatin resistance in an ovarian cancer cell line. Gynecol Oncol 2005;97:26-34. [Crossref] [PubMed]
- Danielsen SA, Eide PW, Nesbakken A, et al. Portrait of the PI3K/AKT pathway in colorectal cancer. Biochim Biophys Acta 2015;1855:104-21. [PubMed]
- Dijkstra JM, Alexander DB. The “NF-ĸB interacting long noncoding RNA”(NKILA) transcript is antisense to cancer-associated gene PMEPA1. F1000Res 2015;4:96. [Crossref] [PubMed]


