
Exopolysaccharides from an Ophiopogon japonicus endophyte inhibit proliferation and migration in MC-4 human gastric cancer cells
Introduction
Gastric cancer is the third leading cause of cancer-associated mortalities worldwide and is a major cause of mortality in Asia (1-3). According to a survey, the mortality rate of gastric cancer was ranked second in overall cancer mortality (4-6). Accordingly, therapeutic strategies for treating gastric cancer are in demand (7-9).
In recent years, natural products have gained increasing attention from a therapeutic point of view and have become promising sources of potential new drugs (10-12). O. japonicus is an evergreen, sod-forming perennial plant that is widely used in traditional Chinese medicine (TCM). The root tuber is the medicinal part of O. japonicus and exerts its antitumor activity by enhancing the immune response of the body against cancer (13-15). Extensive studies have revealed that the major active ingredients in O. japonicus are polysaccharides, which can not only reduce the toxicity caused by chemotherapy drugs, but also possess synergistic antitumor activity (16,17).
An endophyte is an endosymbiont that lives within a healthy plant in its extracellular space or inside the cell. A large number of studies have reported that the medicinal active ingredients produced by endophytes isolated from medicinal plants showed anti-tumor, antibacterial, antiviral and anti-oxidant activities (18-20). Microbial exopolysaccharides (EPSs) are metabolic products of entophytes that accumulate on the microbial cell surface (21). Numerous studies have reported its biological functions, such as protection against environmental stressors and as a component of mucous membranes (22).
In a previous study, our group investigated an entophytic microorganism isolated from O. japonicus and was the first to discover the antitumor exogenous polysaccharide from the entophytic microorganism (23). In this study, we explored the effect of EPSs extracted from the O. japonicus endophyte MD4 on the proliferation and migration of the gastric cancer cell line MC-4.
Methods
Cell culture
The human gastric cancer cell line MC-4 was obtained from the Zhejiang Provincial Center for Disease Control and Prevention (Zhejiang CDC). MC-4 cells were cultured in RPMI-1640 culture medium (Genome Biotechnology, Hangzhou, China) supplemented with 10% (v/v) fetal bovine serum (Zhejiang Tianhang Biotechnology Co., Ltd., Hangzhou, China), 100 U/mL penicillin and 100 µg/mL streptomycin under 37 °C, 5% CO2, and water saturated humidity conditions.
Isolation of the endophytic bacteria MD4 from O. japonicus
The fresh roots of O. japonicus were rinsed with sterile water to remove the sediment from the root surface. Then, the root surface was washed with 75% ethanol for 30 s, and rinsed with sterile water 3 times, followed by 0.1% mercuric chloride for 5 min. The root samples were sectioned into 4–6 mm slabs using a sterile scalpel and then transferred onto lysogeny broth (LB) plates, followed by incubation at 37 °C for 14 days. This length of time allows the growth of endophytic bacteria from the sections. A single bacterial colony was picked from an LB medium plate for-purification. The purifying process was repeated several times. The purified bacteria, temporarily named MD4, were inoculated into 5 mL LB liquid medium in conical flasks maintained at 37 °C and cultured 12 h prior to use.
Identification of the endophytic bacteria MD4
After 12 h cultured, the bacteria in LB liquid medium were separated by centrifugation (12,000 rpm, 1 min). After removing the supernatant, the bacterial pellet was scraped into a 2 mL centrifuge tube, washed with TE buffer (1 mL, 1 M Tris-HCl, 0.5 M EDTA; pH 8.0) and centrifuged (12,000 rpm, 1 min). After discarding the supernatant, the isolated endophytic bacteria were resuspended in 200 µL TE buffer with 50 µL lysozyme and incubated at 37 °C for 60 min with gentle agitation. A total of 10 µL of proteinase K was added to the suspension, followed by incubation at 37 °C for 30 min with gentle agitation. The suspension was then supplemented with 40 µL 10% SDS and incubated at 37 °C for 30 min with gentle agitation. Subsequently, the suspension was mixed with NaCl (100 µL, 5 M) and sodium acetate solutions (200 µL, 3 M). Equal volumes of chloroform and isoamylol were added to the suspension, and the sample was gently shaken for seconds. Following incubation at room temperature for 5 min, the mixture was centrifuged (8,000 rpm, 10 min), and the supernatant was transferred into precooled isopropanol and maintained for 5 min. Following centrifugation (12,000 rpm, 10 min), the deposit was washed with 1 mL 70% ethanol and dried by evaporation at room temperature. The final deposits (dissolved in 30 µL high-salt TE buffer) were collected for the following analysis. After electrophoresis was conducted in a 1% (w/v) agarose gel, the gel was stained with ethidium bromide and visualized with UV. PCR was conducted using the universal primers (forward, 5'-AGAGTTTGATCCTGGCTCAG-3'; reverse, 5'-AAGGAGGTGATCCAGCCGCA-3') to determine the 16S rDNA gene sequence. PCR amplification consisted of an initial denaturation at 94 °C for 5 min followed by 30 cycles (denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min and extension at 72 °C for 2 min) and a final extension at 72 °C for 8 min. Amplified DNA was sequenced by Takara Biotechnology Co., Ltd. The sequence of 16S rDNA was subjected to BLAST analysis with the NCBI database and aligned by using the multiple sequence alignment program, CLUSTAL X. A phylogenetic analysis was performed using MEGA 5.0. The evolutionary history was inferred using the neighbor-joining method.
EPSs extraction
The endophyte MD4 was seeded in beef extract peptone medium (3.0 g beef extract, 10.0 g peptone and 5.0 g NaCl dissolved in 1,000 mL) and incubated at 37 °C for 48 h while shaking, to increase the bacterial concentration. Following centrifugation (3,600 rpm, 30 min), the supernatant of the bacterial suspension was treated with activated carbon (Huadong Medicine Co. Ltd., Hangzhou, China) to bleach the medium and incubated at 40 °C for 30 min. After the removal of the activated carbon, the supernatant was concentrated by rotary evaporation, then the Sevag (4:1 ratio of chloroform to normal butanol) method was used to dislodge the proteins. EPSs were extracted by ethanol precipitation, and the final precipitate was dissolved in PBS and then dialyzed with PEG 20000 (Sinopharm Chemical Reagent Co. Ltd., Shanghai, China) to obtain the desired MD4-EPS for our further experiments. The total carbohydrate content, as well as MD4-EPS concentrations in the precipitated EPSs, was evaluated using the phenol-sulfuric acid method.
MTT assay
The MTT test was used to assay cell proliferation. The succinate dehydrogenase in the mitochondria of living cells reduces the exogenous MTT to the water-insoluble blue-purple crystalline formazan and deposits in the cells, whereas dead cells have no such function. Within a certain number of cells, the amount of crystallization of MTT was proportional to the number of cells. MC-4 cells (2×105 cells /mL) were seeded onto a 96-well plate and incubated for 24 h. Subsequently, the cells were treated with different concentrations of MD4-EPS (0.84, 1.26, 1.68, 2.10, 2.52, 2.94 and 3.36 µg/µL) and incubated at 37 °C with 5% CO2 for 24 h. MTT solution (20 µL) was added to each well, the samples were and incubated for 4 h. The optical density (OD) was measured at a wavelength of 492 nm in a Labsystems Multimode Reader (Labsystems, Finland), and the viability rate was calculated from three independent biological replicates. The control group was used as the reference group with cell viability set at 100%.
Wound healing assay
MC-4 cells (2×105 cells/well) were seeded onto 12-well plates. The cells were cultured under standard conditions for approximately 24 h. When the cells reached a confluence of approximately 80%, scratch wounds were made by scraping a line with a sterile 200 µL pipette tip, and then the cells were washed 3 times with PBS. The cells were then treated with three different concentrations of MD4-EPS (0.84, 1.68 and 3.36 µg/µL) and incubated at 37 °C and 5% CO2 for 24 h to allow the migration into the cell-free areas from the surrounding monolayer. After incubation for 24 h, the wound closure was photographed with a light microscope at 40× magnification. Relative quantitation of the cell migration index was quantified using Image-Pro Plus 6.0 analysis software.
Gelatin zymography assay
MMP2 and MMP9 enzymatic activities were analyzed using gelatin zymography. MC-4 cells (2×105 cells/well) were plated in 6-well plates and treated with different concentrations of MD4-EPS (0.84, 1.68 and 3.36 µg/µL) in the wound healing assay. After incubation for 24 h, the culture media were collected and centrifuged (1,000 rpm, 10 min) at 4 °C to remove cell debris. The supernatant was collected and mixed with sample loading buffer (50 mM Tris-HCl pH 6.8, 10% glycerol, 1% SDS, 0.01% bromophenol blue) without DTT to denature MMPs. The samples were loaded onto a 7.5% SDS-polyacrylamide gel (containing 0.1% gelatin). Then, the gel was soaked twice for 30 min in 2.5% Triton-X-100 to remove SDS and transferred into the incubation buffer (50 mM Tris-HCl pH 7.5, 5 mM CaCl2, 1 µM ZnCl2 and 0.05% NaN3) at 37 °C for 18–20 h. The gel was immersed in staining solution (0.05% Coomassie blue RR-250, 30% methanol and 10% acetic acid in dH2O) for 3 h and subsequently in destaining solution (5% methanol and 10% acetic acid in dH2O) for 30 min. After the action of MMP2 and MMP9, the gel was decomposed, and the sites affected by the enzyme showed transparent bands. The visualized bands indicated the presence of MMPs with gelatinolytic activity. The comparison with standard protein marker bands indicated that MMP2 was detected by the clear band at 72 kDa and MMP9 at 92 kDa.
Enzyme-linked immunosorbent assay (ELISA)
MC-4 cells were seeded as previously described in the wound healing assay and treated with different concentrations of MD4-EPS for 24 h. After incubation, the cell culture medium was collected and centrifuged (1,000 rpm, 10 min) to remove the cells. The supernatant was transferred to new tubes and stored at −80 °C. MMP9 and MMP2 ELISA kits (ExCell Biology, Inc., Shanghai, China) were used to detect MMP9 and MMP2 concentrations in the cell culture supernatants, according to the manufacturer’s protocol. The OD was measured at 450 nm in the Labsystems Multimode Reader (Labsystems, Finland), and each group was analyzed in triplicate. MMP9 and MMP2 concentrations were determined by a reference curve plotted using the kit-provided standards.
Statement of ethics approval
This manuscript does not need any ethics approval because the experimental materials are MC-4 cells.
Statistical analysis
All experiments were performed in triplicate. Data are shown as the mean ± standard deviation (SD). The Windows-based SPSS software (version 16.0) was used for statistical analysis. The MTS cell viability, migration rate, enzymatic activity and protein secretion level data were subjected to one-way analysis of variance (ANOVA), followed by Fisher’s least significant difference (LSD) post hoc test. P<0.05 indicated a statistically significant difference.
Results
Isolation and identification of endophytic bacteria from O. japonicus
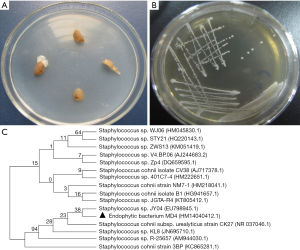
As shown in Figure 1A, endophytic bacteria were isolated from slices of the tuberous roots of O. japonicus on LB plates. A single bacterial colony was picked from LB medium plates for purification (Figure 1B). A gram-positive bacterium, named MD4, with sticky colony characteristics was isolated from the inner section of O. japonicus and confirmed as Staphylococcus sp. A partial sequence of the 16S rDNA gene was identified and deposited in GenBank at NCBI (Figure 1, Accession No. HM140412.1). Evolutionary analyses were conducted in MEGA5. As shown in Figure 1C, BLAST analysis revealed that MD4 demonstrated the highest similarity (99%) with-Staphylococcus sp. A neighbor-joining dendrogram was constructed for the phylogenetic analysis of the endophyte bacterium MD4. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown above the branches. The evolutionary distances were computed using the maximum composite likelihood method and are reported as the number of base substitutions per site.
Concentrations of EPSs extracted from the endophyte MD4
Using the phenol-sulfuric acid method, MD4-EPS concentrations were obtained by a standard curve using a glucose solution. The results of the phenol sulfuric acid method showed that the purified EPS concentration from MD4 was 28.068 µg/µL.
Effects of MD4-EPS on MC-4 cell proliferation
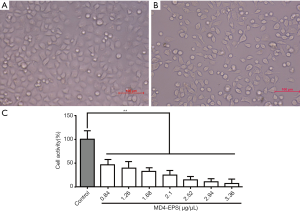
Cell viability was evaluated by MTT assay after 24 h of treatment with EPS. The statistical analysis of cell viability is shown in Figure 2A,B,C. When MC-4 cells were treated for 24 h with graded concentrations of MD4-EPS (0.84, 1.26, 1.68, 2.10, 2.52, 2.94 and 3.36 µg/µL), the cells decreased in size and presented a spherical appearance. Besides, MC-4 cell viability gradually decreased with increasing MD4-EPS concentration. In addition, the IC50 value of the MD4-EPS was 0.891 µg/µL. Therefore, the results of MTT assay showed that MC-4 cells treated with MD4-EPS inhibited cell proliferation in a concentration-dependent manner.
Effect of MD4-EPS on migration
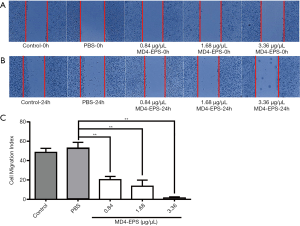
The effect of the EPSs extracted from the endophyte MD4 on the migration of MC-4 cells was quantified using the cell wound healing assay. The effect of MD4-EPS on cell migration was assessed with wound margin analysis (Figure 3A,B). As shown in Figure 3C, at high EPS concentrations, the cells became round, and some of them were shed and suspended in culture medium. Microscopic analysis indicated that MD4-EPS decreased the migration rate of MC-4 cells in a dose-dependent manner compared with normal cells and cells in the PBS negative control group. The migration of MC-4 cells was significantly inhibited at the MD4-EPS concentration of 1.68 µg/µL, whereas the cells of the control group and the negative control PBS group did not show any inhibition in their migration ability. It can be seen from the results of the wound healing assays that EPSs exhibited a strong ability to inhibit migration. When EPSs were added to the cells, they inhibited cell migration, while the cells in the control group and the PBS group maintained their original migration ability.
Effect of MD4-EPS on MMP2 and MMP9 enzymatic activities
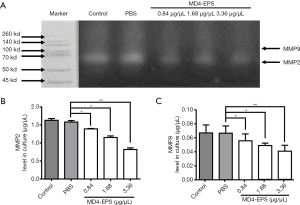
The gelatin zymography analysis results showed that the migration-related enzymatic activities of MMP2 and MMP9 were decreased after EPS treatment. As shown in Figure 4A, the results indicated that MMP2 enzymatic activity decreased as the concentration of MD4-EPS increased. MMP9 also showed decreased enzymatic activity due to MD4-EPS treatment, but the inhibitory effect on MMP9 was stronger than the effect on MMP2.
Effect of MD4-EPS on MMP2 and MMP9 protein secretion levels
ELISA assays evaluated MMP2 (Figure 4B) and MMP9 (Figure 4C) protein concentrations in the cell culture medium after MD4-EPS treatment. Our results showed that MMP2 and MMP9 protein levels were gradually reduced in a concentration-dependent manner with the increasing concentration of MD4-EPS compared with the PBS group. Moreover, there were no significant differences between the control group and the PBS group.
Discussion
In the present study, we explored the antitumor effect of EPSs extracted from the endophyte MD4, which was identified as Staphylococcus sp. based on 99% similarity in the partial sequence analysis of 16S rDNA. According to the most recent publications, the antitumor ability of EPSs from endophytes is mainly due to the effect of EPS on the immune system and on the induction of tumor cells apoptosis (24).
The main biological characteristics of tumor cells are uncontrolled proliferation and high migration ability (25,26). The proliferation of MC-4 cells was reduced in a concentration-dependent manner by MD4-EPS. According to the results of wound healing assays, EPSs exhibited strong abilities to inhibit migration. When EPSs were added to the cells, they inhibited cell migration, while the cells in the control and PBS groups maintained their original migration ability. These results suggested that MD4-EPS significantly reduced MC-4 cell proliferation and further affected migration.
MMPs degrade collagen type IV in the basement membrane, and they have a close relation with malignant tumor infiltration and migration (27-29). In this study, the antitumor effect of EPSs was exerted by the inhibition of MMP2 and MMP9 protein activation and secretion. Indeed, the results of the gelatin zymography assay showed that MMP2 and MMP9 enzymatic activities were suppressed by the gradual increase in MD4-EPS concentration. ELISA showed the same trend in the secretion of MMP2 and MMP9 protein into the basement membrane after treatment with MD4-EPS. It seems that the MD4-EPS inhibition of MC-4 cell proliferation was mediated by decreasing the secretion of MMP2 and MMP9. In other words, MC-4 cell migration was inhibited by EPSs that were also linked to the downregulation of MMP2 and MMP9. Furthermore, MMP2 and MMP9, two important members of the MMP family-involved in the degradation of the extracellular matrix, exist in various bodily tissues (30).
In this study, we found that the enzymatic activities of MMP2 and MMP9 protein were significantly suppressed by MD4-EPS. This result can be explained by the abnormally high levels of MMP2 and MMP9 in gastric cancer cells that increase the degradation of the ECM and break the basal membrane, thereby leading to tumor migration (31,32). Our results also provide evidence for additional treatments targeting the pathogenesis of gastric cancer migration and for the development of drugs that target MMP2 and MMP9.
Cell proliferation is a normal physiological process occurring in every tissue of the body (33). Under normal conditions, cell proliferation and programmed cell death exist in a balance (34). However, in gastric cancer cells, this balance is disrupted by uncontrolled cell proliferation (35). Therefore, inhibiting cell proliferation is considered to be an effective approach to cancer therapy. Numerous signaling pathways have been reported to be involved in the cell proliferation of cancer cells, such as the Wnt, JAK/STAT, PI3K/Akt and MAPK signaling pathways (36,37). The MAPK signaling pathway is the key signaling pathway in gastric cancer cells, and it manipulates cell proliferation and migration by regulating MMPs or other proteins (38). In this study, MMPs in MC-4 human gastric cancer cells were significantly downregulated by MD4-EPS treatment, suggesting the potential effects of MD4-EPS on the MAPK signaling pathway, which will be studied in the future.
Conclusions
In summary, EPSs clearly showed an antitumor effect via MD4-EPS treatment by inhibiting cell proliferation and migration.
Acknowledgments
Funding: The present study was supported by grants from the National Natural Science Foundation Project (No. 81874355) and the Zhejiang Provincial Natural Science Foundation (No. LY18H280008). The present study was also supported by the Subject of Zhejiang TCM Administration Bureau (No. 2006C019).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2018.11.28). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This manuscript does not need any ethics approval because the experimental materials are MC-4 cells. Informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Han P, Wang QL, Zhang X. Expression of TRAP1 in gastric cancer tissue and its correlation with malignant biology. Asian Pac J Trop Med 2016;9:67-71. [Crossref] [PubMed]
- Choi HJ, Ki CS, Suh SP, et al. Presymptomatic identification of CDH1 germline mutation in a healthy korean individual with family history of gastric cancer. Ann Lab Med 2014;34:386-9. [Crossref] [PubMed]
- He J. Strengthen the cancer surveillance to promote cancer prevention and control in China. Zhonghua Zhong Liu Za Zhi 2018;40:1-4. [PubMed]
- Oweira H, Schmidt J, Mehrabi A, et al. Comparison of three prognostic models for predicting cancer-specific survival among patients with gastrointestinal stromal tumors. Future Oncol 2018;14:379-89. [Crossref] [PubMed]
- Tehfe M, Tabchi S, Laterza MM, et al. Ramucirumab in HER-2-positive gastroesophageal adenocarcinoma: an argument for overcoming trastuzumab resistance. Future Oncol 2018;14:223-8. [Crossref] [PubMed]
- Wei J, Yang Y, Du J, et al. Peritoneal metastasis in relation to outcome and therapeutic strategy in gastric cancer. Transl Cancer Res 2017;6:149-56. [Crossref]
- Lyu S, Ye L, Li T, et al. Liver resection of metastases for colorectal cancer, gastric cancer and breast cancer: two hospital experiences. Transl Cancer Res 2016;5:562-9. [Crossref]
- Li L, Cheng X, Zheng Z, et al. Overexpression of cancer cell-derived immunoglobulin G correlates with poor prognosis in gastric cancer patients. Transl Cancer Res 2016;5:285-93. [Crossref]
- Chen MH, Chen XJ, Wang M, et al. Ophiopogon japonicus--A phytochemical, ethnomedicinal and pharmacological review. J Ethnopharmacol 2016;181:193-213. [Crossref] [PubMed]
- Zhang Y, Han Y, Zhai K, et al. Ophiopogonin-D suppresses MDA-MB-435 cell adhesion and invasion by inhibiting matrix metalloproteinase-9. Mol Med Rep 2015;12:1493-8. [Crossref] [PubMed]
- Wang Y, Yan T, Shen J, et al. Preventive effect of Ophiopogon japonicus polysaccharides on an autoallergic mouse model for Sjogren's syndrome by regulating the Th1/Th2 cytokine imbalance. J Ethnopharmacol 2007;114:246-53. [Crossref] [PubMed]
- Agnes A, Lirosi MC, Panunzi S, et al. The prognostic role of perioperative allogeneic blood transfusions in gastric cancer patients undergoing curative resection: A systematic review and meta-analysis of non-randomized, adjusted studies. Eur J Surg Oncol 2018;44:404-19. [Crossref] [PubMed]
- Fang J, Wang X, Lu M, et al. Recent advances in polysaccharides from Ophiopogon japonicus and Liriope spicata var. prolifera. Int J Biol Macromol 2018;114:1257-66. [Crossref] [PubMed]
- Auyeung KK, Han QB, Ko JK. Astragalus membranaceus: A Review of its Protection Against Inflammation and Gastrointestinal Cancers. Am J Chin Med 2016;44:1-22. [Crossref] [PubMed]
- Gong Y, Zhang J, Gao F, et al. Structure features and in vitro hypoglycemic activities of polysaccharides from different species of Maidong. Carbohydr Polym 2017;173:215-22. [Crossref] [PubMed]
- Kumaran RS, Kim HJ, Hur BK. Taxol-producing [corrected] fungal endophyte, Pestalotiopsis species isolated from Taxus cuspidata. J Biosci Bioeng 2010;110:541-6. [Crossref] [PubMed]
- Chan JK, Brady MF, Penson RT, et al. Weekly vs. Every-3-Week Paclitaxel and Carboplatin for Ovarian Cancer. N Engl J Med 2016;374:738-48. [Crossref] [PubMed]
- Moscovici M. Present and future medical applications of microbial exopolysaccharides. Front Microbiol 2015;6:1012. [Crossref] [PubMed]
- da Silva WJ, Goncalves LM, Seneviratne J, et al. Exopolysaccharide matrix of developed Candida albicans biofilms after exposure to antifungal agents. Braz Dent J 2012;23:716-22. [Crossref] [PubMed]
- Moreira Â, Pereira SS, Costa M, et al. Adipocyte secreted factors enhance aggressiveness of prostate carcinoma cells. PLoS One 2015;10:e0123217 [Crossref] [PubMed]
- Ou Y, Xu S, Zhu D, et al. Molecular mechanisms of exopolysaccharide from Aphanothece halaphytica (EPSAH) induced apoptosis in HeLa cells. PLoS One 2014;9:e87223 [Crossref] [PubMed]
- Chen YT, Yuan Q, Shan LT, et al. Antitumor activity of bacterial exopolysaccharides from the endophyte Bacillus amyloliquefaciens sp. isolated from Ophiopogon japonicus. Oncol Lett 2013;5:1787-92. [Crossref] [PubMed]
- Chang J, Zhang Y, Li Y, et al. NrF2/ARE and NF-kappaB pathway regulation may be the mechanism for lutein inhibition of human breast cancer cell. Future Oncol 2018;14:719-26. [Crossref] [PubMed]
- Wei HC. A mathematical model of tumour growth with Beddington-DeAngelis functional response: a case of cancer without disease. J Biol Dyn 2018;12:194-210. [Crossref] [PubMed]
- Hu K, Zhang J, Yu M, et al. Inhibition of Mnk-eIF4E pathway sensitizes the efficacy to chemotherapy in anaplastic thyroid cancer. Future Oncol 2017;13:489-98. [Crossref] [PubMed]
- Gallop-Evans E. The role of alisertib in treatment of peripheral T-cell lymphomas. Future Oncol 2015;11:2515-24. [Crossref] [PubMed]
- Tatti O, Gucciardo E, Pekkonen P, et al. MMP16 Mediates a Proteolytic Switch to Promote Cell-Cell Adhesion, Collagen Alignment, and Lymphatic Invasion in Melanoma. Cancer Res 2015;75:2083-94. [Crossref] [PubMed]
- Radunovic M, Nikolic N, Milenkovic S, et al. The MMP2 and MMP9 promoter polymorphisms and susceptibility to salivary gland cancer. J BUON 2016;21:597-602. [PubMed]
- Lu S, Zhang Z, Chen M, et al. Silibinin inhibits the migration and invasion of human gastric cancer SGC7901 cells by downregulating MMP2 and MMP9 expression via the p38MAPK signaling pathway. Oncol Lett 2017;14:7577-82. [PubMed]
- Xi Q, Qi H, Li Y, et al. Low frequency ultrasound combined with baicalein can reduced the invasive capacity of breast cancer cells by down regulating the expression of MMP-2, MMP-9, and u-PA. Transl Cancer Res 2016;5:838-44. [Crossref]
- Duan L, Wu Y, Liang Y, et al. MiRNA-516b inhibits ameloblastoma cell proliferation and invasion by regulating MYCBP/c-myc/RECK/MMP pathway. Transl Cancer Res 2017;6:667-78. [Crossref]
- Chen B, Li Q, Zhou Y, et al. The long coding RNA AFAP1-AS1 promotes tumor cell growth and invasion in pancreatic cancer through upregulating the IGF1R oncogene via sequestration of miR-133a. Cell Cycle 2018;17:1949-66. [Crossref] [PubMed]
- Yang Z, Jiang X, Li D, et al. TCF21 inhibits proliferation and chemoresistance through the AKT pathway in human gastric cancer. Gene 2019;682:42-9. [Crossref] [PubMed]
- Cao G, Chen D, Liu G, et al. CPEB4 promotes growth and metastasis of gastric cancer cells via ZEB1-mediated epithelial-mesenchymal transition. Onco Targets Ther 2018;11:6153-65. [Crossref] [PubMed]
- Gao X, Sun J, Huang C, et al. RNAi-mediated silencing of NOX4 inhibited the invasion of gastric cancer cells through JAK2/STAT3 signaling. Am J Transl Res 2017;9:4440-9. [PubMed]
- He Y, Ge Y, Jiang M, et al. MiR-592 Promotes Gastric Cancer Proliferation, Migration, and Invasion Through the PI3K/AKT and MAPK/ERK Signaling Pathways by Targeting Spry2. Cell Physiol Biochem 2018;47:1465-81. [Crossref] [PubMed]
- Kim S, Choi MG, Lee HS, et al. Silibinin suppresses TNF-alpha-induced MMP-9 expression in gastric cancer cells through inhibition of the MAPK pathway. Molecules 2009;14:4300-11. [Crossref] [PubMed]


