
Circular RNA hsa_circ_0002124 promotes hepatocellular carcinoma cell proliferation through the MAPK pathway
Introduction
Globally, hepatocellular carcinoma (HCC) accounts for approximately 90% of all primary liver cancers. HCC is one of the leading causes of cancer-related deaths, of which 50% occur in China (1). Currently, the most effective treatment for HCC remains surgical treatment. However, because of the lack of effective strategies for early diagnosis, only 30–40% of patients with HCC are considered suitable for radical resection. Nevertheless, because of the high recurrence and metastasis rate following surgery, the prognosis of these patients with HCC remains poor (2). Therefore, more effective tumor markers are the key to early diagnosis, early treatment, and improved prognosis of HCC.
Circular RNA (circRNA) is a type of noncoding RNA that is widely detected in mammals. circRNA is primarily involved in gene regulation in vivo (3-5). Most of the circRNAs are derived from the exon region of the gene, whereas a small part is formed by intron splicing (6,7). Unlike long noncoding RNAs (lncRNAs) and microRNAs (miRNAs), circRNA does not contain a 5’ end and a 3’ end structure; they are formed by a covalently closed cyclic structure (8). CircRNA is widely involved in human physiological and pathological regulation: (I) circRNA can act as a miRNA “spill” (miRNA sponge); (II) circRNA could interact with target proteins, and (III) circRNA can be translated into peptides (9). Studies on cancer have reported that circRNA acts as “miRNA cavernous body” and could indirectly regulate downstream target genes (10,11). It has been reported that one circRNA contains at least one miRNA binding site. Moreover, circRNAs can adsorb miRNA as a “cavernous body” of RNA. Therefore, circRNAs could regulate gene expressions through the mechanism of competitive endogenous RNAs (ceRNAs) (9). In 2010, Pandolfi et al. have proposed that lncRNAs, pseudogene RNA, and circRNA contain a common site that can be bound by miRNAs (12). The competitive binding of miRNAs leads to the formation of a mutually regulated network between these different types of RNA that plays an important role in the various cell physiological processes (13). In HCC, a previous study has suggested that circHIPK3 can act as a “cavernous body” to adsorb miR-124. Therefore, IL6R and DXL2 expressions were upregulated via the ceRNA mechanism, thereby promoting the proliferation of HCC cells (14). Although the current studies have suggested the importance of its role in liver cancer, additional number of circRNAs and their detailed functions in HCC remains to be evaluated in the future.
Hsa_circ_0002124, first reported by Jeck et al. in 2013, is located in chr15:41667909-41669502, which is the region of intron 9 in NuSAP1 (15). It is reported that NUSAP1 may be a target of miR-122 involved in cell cycle-associated processes during HCC progression (16). Moreover, the increased NUSAP1 expression had been detected in HCC cells and tissues, and increased levels of NUSAP1 in HCC samples have been correlated with shorter survival time of patients (17). These evidences implicate the role of NUSAP1 in HCC progression. As the hsa_circ_0002124 expression and functions in HCC remain largely unknown, further exploration on this topic could provide insights into the understanding of circRNAs as a whole in the carcinogenesis of HCC. A literature review showed that hsa_circ_0002124 has not previously been reported to be directly involved in HCC; to the best of our knowledge, this is the first study to investigate the role of hsa_circ_0002124 in HCC progression. In the present study, we employed PCR to identify hsa_circ_0002124 in HCC cells. The hsa_circ_0002124 expression in clinical samples was tested using qPCR. Moreover, the cell functions of hsa_circ_0002124 in HCC cells were examined with the knockdown and overexpressed hsa_circ_0002124 in 97H cells. Gene ontology (GO) and KEGG analysis were further performed to uncover the key functions and pathways in HCC. The interaction networks between hsa_circ_0002124 and its downstream miRNAs and genes were constructed using the Cytoscape software. Additionally, Western blotting was used to confirm the expression changes of key proteins (i.e., p-JNK, p-ERK, p-P38, and c-Myc) in the MAPK pathway. Our results would be helpful to understand the detailed role of hsa_circ_0002124 in HCC cells.
Methods
Clinical samples
The present experiment was approved by the Committee on the Ethics of Sun Yat-sen University (approval No. SYSU-20161078). All subjects were completely informed about the study goals and procedure and provided their signed informed consent. The HCC tissues and their paired pericarcinomatous tissues of 20 patients with HCC were collected according to the standard operation procedures at a third affiliated hospital of the Sun Yat-sen University. The biopsies were immediately frozen and stored at −80 °C until further processing for total RNA preparation.
Cell treatment
The LO2 cells, HepG2 cells, Huh7 cells, 97L cells, 97H cells, Hep3B cells, and LM3 were obtained from ATCC (Virginia, USA) and maintained in RPMI 1640 with 10% (v/v) FBS (Invitrogen, USA). The culture medium contained 100 U/mL penicillin/streptomycin. The cells were cultured in a humidified chamber with 5% CO2 at 37 °C. After 3 days, 0.25% of trypsin (Thermo Fisher Scientific, USA) was routinely used for digestion. The log phase cells with excellent growth conditions were selected for further experimentation. Sequence hsa_circ_0002124 was downloaded from the cirBase database (http://www.mirbase.org/). The circRNA overexpression vector PCDNA 3.1-circRNA was purchased from Addgene (USA). In the present study, we selected 97H cells to perform cell transfection. The QIAGEN Plasmid Mini Kit (QIAGEN, Germany) was used to obtain purified plasmids according to the manufacturer’s protocol. Subsequently, PCDNA 3.1-circRNA was transfected into 97H cells via the Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific, Waltham, MA, USA). The PCDNA 3.1-circRNAs were added to the transfection reagents diluted in the Opti-MEM medium. The mixture was incubated at room temperature for 10–15 min to form the PCDNA 3.1-circRNA–transfection reagent complexes, followed by its addition to the cells and further incubation for 48 h. Moreover, double-stranded siRNAs (dsRNA) targeting hsa_circ_0002124 were synthesized by chemical methods (ReiBo Biotech, China). The nucleotide sequence of the dsRNA for hsa_circ_0002124 was 5'-CUCCAGACAAAUUAUUACC-3' and 3'-GAGGUCUGUUUAAUAAUGG-5'. The nucleotide sequence of the control siRNA from a scramble sequence was 5'-UUCUCCGAACGUGUCACGUTT-3' and 3'-TTAAGAGGCUUGCACUGAGCA-5'. The cells were seeded at 5×105 cells per well in 6-well plates in DMEM containing 10% FBS without penicillin and streptomycin and incubated overnight. Transfection experiments were performed with the OPTI-MEM serum-free medium and Lipofectamine 2000 reagent at a final siRNA concentration of 100 nM. qRT-PCR was performed to validate the effectiveness of the treatments.
qPCR
The TaKaRa MiniBEST Universal Genomic DNA Extraction Kit (TaKaRa, Japan) was applied to harvest the gDNA of HCC tissues and paired pericarcinomatous tissues. The detailed operation products were obtained according to the manufacturer’s instructions. Moreover, the TaKaRa MiniBEST Universal RNA Extraction Kit (TaKaRa, Japan) was used to exact total RNAs from LO2 cells, HepG2 cells, Huh7 cells, 97L cells, 97H cells, Hep3B cells, LM3, cell mixture of 7 cells (1×106 cells from each cell line), and tissue samples. cDNAs of different samples were synthesized by the PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa, Japan). The experiment was conducted as per the manufacturer’s instructions. The concentration of gDNA and cDNA were assessed using NanoDrop 6000 (Thermo Fisher, USA). The OD260/OD280 value of 1.8–2.1 was considered to represent an ideal sample. Furthermore, the integrity of gDNA and cDNA was examined on denaturing agarose gel. We designed the following two primer pairs to identify the liner DNA and the corresponding circRNA (hsa_circ_0002124): circRNA (120 bp): hsa_circ_0002124-CF, 5'-AAACAACCCCATCTCCAGACA-3' and hsa_circ_0002124-CR, 5'-AGGGGACGAGACAAACTTGC-3'; liner DNA (123 bp): hsa_circ_0002124-LF, 5'-GTTGACAACTGAGGCAACGC-3' and hsa_circ_0002124-LR, 5'-TGCCCCCATGGTTTTAGCTT-3'. PCR reaction system (20 µL) was used in the present study constituting 10-µL 2× PCR Taq MasterMix with dye (Applied Biological Materials, USA), 0.5-µL primer mixture (5 pmol each), 1-µL DNA template (approximately 20 ng), and 8.5-µL ddH2O under the following thermal conditions: 2 min at 95 °C, followed by 40 cycles at 95 °C for 1 min, 60 °C for 1 min, 72 °C for 1 min, and 72 °C for 7 min. The PCR products were analyzed using agarose gel electrophoresis (1.5%) and Sanger sequencing. Moreover, we performed qPCR to determine the hsa_circ_0002124 expression. GAPDH was employed as an internal gene. The 10-µL qPCR system contained 5-µL 2× SsoAdvanced™ Universal SYBR® Green Supermix (BioRad, USA), 0.5-µL primers (5 pmol each), 0.5-µL DNA template (approximately 20 ng), and 4-µL ddH2O. The QuantStudio 6 Flex Real-Time PCR System (ABI, USA) was used to conduct the following thermal steps: 3 min at 98 °C, followed by 40 cycles at 95 °C for 15 s and at 60 °C for 25 s. The ΔΔct method was used to calculate the RNA expression with the relative standard curve.
GO and KEGG analysis
Hsa_circ_0002124 were uploaded into the function annotation portal of DAVID database, as described previously (18). Following the enrichment annotation, a list of GO items were obtained. The related pathways of the parental genes of has_circ_0002124 were analyzed using KEGG. P<0.05 and FDR <0.05 were considered significant.
Interaction analysis
Cytoscape is an online bioinformatics resource platform for constructing biomolecular interaction networks (18). To investigate the ceRNA mechanism of hsa_circ_0002124, the target miRNAs and genes were analyzed using Cytoscape platform to identify the densely connected region in the PPI network from the STRING database (19).
Cell proliferation, behavior, cycle, and apoptosis
Cell proliferation of differently-treated cells was tested using the MTT Kit (Sigma Aldrich, USA). The OD values were collected at three different time points (24, 48, and 72 h). The Oris™ Cell Migration Assay (Amsbio, UK) was used to examine cell migration. Briefly, 1.5×105 mock-vehicle-treated 97H cells and 1.5×105 overexpressed hsa_circ_0002124-treated 97H cells were seeded on the top chamber of each transwell. After culturing at 37 °C for 24 h, the cells that passed through the bottom side membrane were fixed with methanol. Hematoxylin was used to stain cells. The cell number was counted under optical microscope. Further, the Cultrex® Cell Invasion Assay (Amsbio, UK) was used to analyze cell invasion by different treatments, according to the manufacturer’s protocol. Furthermore, we analyzed the cell cycle of differently-treated 97H cells. Flow cytometry was applied to analyze the cell cycle distribution of mock-vehicle-treated 97H cells and overexpressed hsa_circ_0002124-treated 97H cells. After 36 h of transfection, the cells were digested with 0.25% trypsin and fixed with 70% ethanol. FACS calibur flow cytometer (BD Biosciences, USA) was employed to test the cell cycle distribution by different treatments. In addition, we analyzed the cell apoptosis of mock-vehicle-treated 97H cells and 1.5×105 overexpressed hsa_circ_0002124-treated 97H cells using the Annexin V-FITC Apoptosis Staining/Detection Kit (Abcam, USA) according to the manufacturer’s protocol.
Western blotting
Mock-vehicle-treated 97H cells (2.9×106) and overexpressed hsa_circ_0002124-treated 97H cells were treated with pre-cooled cell lysate (volume ratio of PMSF: RIPA cell lysate =1:120). The slurry was centrifuged with 13,000 r/min for 5 min (in a low-temperature mode). Subsequently, the supernatant was transferred to a new 1.5-mL tube, which was immediately stored at −80 °C. The protein loading buffer was added to the collected protein samples. The mixture was heated on a 100 °C boiling water bath for 5 min to completely denature the protein. Thereafter, these proteins were transferred onto a polyvinylidene difluoride membrane (Millipore, USA). After being blocked for 1 h at room temperature, the membrane was incubated with rabbit polyclonal anti-mouse p-JNK (1:2,000), JNK (1:2,000), p-ERK (1:2,000), ERK (1:2,000), p-P38 (1:2,000), P38 (1:2,000), c-Myc (1:2,000), and β-Actin (1:2,000) antibody (BOSTER, USA) for 12 h. Subsequently, the treated proteins were incubated with the corresponding secondary antibody (1:2,000 dilution) for 1 h at room temperature (BOSTER, USA). An ECL chemiluminescence detection kit was used to stain the protein bands (Beyotime Institute of Biotechnology, China), and the Odyssey Infrared Imaging System (LI-COR Biosciences, USA) was used to observe and quantify the protein bands.
Statistics
Data were plotted and assessed using Graphad Prism software and expressed as mean ± standard deviation (mean ± SD). Two independent sample t-tests and two-way ANOVA were used in the experiment, and the difference obtained was considered significant at P<0.05.
Results
Hsa_circ_0002124 could be identified in HCC cells
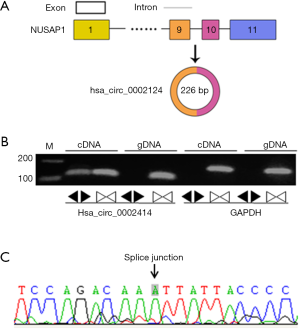
Hsa_circ_0002124 was first reported by Jeck et al. in 2013 (15). Figure 1A suggested the structure of hsa_circ_0002124 to be 226 bp in length and derived from the intron 9 of NUSAP1. To identify hsa_circ_0002124 in HCC cells, we designed convergent and divergent primers to perform PCR reaction using cDNA and gDNA of a mixture of liver cells (Figure 1B). The results indicated that PCR production of hsa_circ_0002124 was obtained with the expected band size from cDNA but not from gDNA. To confirm the results, we performed Sanger sequencing using the cDNA PCR product (Figure 1C), following which the back-spliced junction site of hsa_circ_0002124 was validated. Therefore, hsa_circ_0002124 could be identified in HCC cells.
The hsa_circ_0002124 expression was upregulated in HCC cells and clinical samples
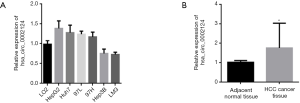
To study the hsa_circ_0002124 expression, we analyzed the expression conditions in HCC tissues, paired pericarcinomatous tissues, and seven liver cells (LO2 cells, HepG2 cells, Huh7 cells, 97L cells, 97H cells, Hep3B cells, and LM3 cells). The hsa_circ_0002124 expression in Hep3B cells and LM3 cells tended to be lower than that in normal liver cells (LO2 cells; Figure 2A). However, the hsa_circ_0002124 expression in HepG2 cells, Huh7 cells, 97L cells, and 97H cells was higher than that in the normal liver cells (LO2 cells). These results suggested the heterogeneity of the hsa_circ_0002124 expressions in different HCC cells. Further, we analyzed the hsa_circ_0002124 expression in HCC tissues of 20 patients and their paired pericarcinomatous tissues (Figure 2B). Our results showed that the hsa_circ_0002124 expressions in HCC tissues were significantly higher than those in paired pericarcinomatous tissues (P<0.05).
Knockdown and overexpressed hsa_circ_0002124 affect 97H cell function
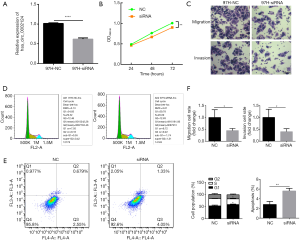
Based on the evidences accumulated in the present study, we speculated that the knockdown and overexpression of hsa_circ_0002124 in 97L cells would affect the cell functions, such as cell proliferation, behavior, cycle, and apoptosis. We first performed that the knockdown of hsa_circ_0002124 in 97H cells could inhibit the cell functions (Figure 3). Figure 3A illustrates the hsa_circ_0002124 expressions in normal- and knockdown-treated 97L cells. The hsa_circ_0002124 expression was significantly inhibited in the knockdown group (P<0.0001). Cell proliferation analysis of 97H cells with or without knockdown hsa_circ_0002124 at 3 time points (i.e., 24, 48, and 72 h) suggested no differences in the OD values obtained at 24 and 48 h, and at 72 h; the OD value of normal 97H cells was significantly higher than that of knockdown hsa_circ_0002124-treated 97H cells (P<0.01) (Figure 3B). Moreover, the analysis of cell migration and invasion of 97H cells without or with the knockdown hsa_circ_0002124 treatments suggested that the cell number of cell migration and invasion were significantly lower in knockdown hsa_circ_0002124-treated 97H cells than in normal 97H cells (P<0.05) (Figure 3C,F). Furthermore, the cell cycle distribution analyses indicated that the cell number of knockdown hsa_circ_0002124-treated 97L cells in the G1 phase was significantly increased and the cell number in the S phase was significantly decreased (P<0.05) compared with that of normal 97L cells, which suggests that the cell cycle was blocked at the G1 phase (Figure 3D,F). In addition, the analysis of the cell apoptosis of 97H cells with knockdown treatment indicated that knockdown hsa_circ_0002124-treated 97H cells significantly promoted cell apoptosis compared with the normal 97H cells (P<0.01, Figure 3E,F). These results revealed that hsa_circ_0002124 could facilitate HCC development.
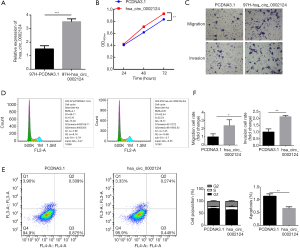
In contrast, we investigated the effectiveness of overexpressed hsa_circ_0002124 in 97L cells (Figure 4). Figure 4A shows the hsa_circ_0002124 expressions in normal- and overexpression-treated 97L cells. The results revealed that the hsa_circ_0002124 expression was significantly promoted in the overexpressed group (P<0.001). Cell proliferation analysis of 97H cells with mock vector-treated or with overexpressed hsa_circ_0002124 at 3 time points (24, 48, and 72 h) showed no differences among the OD values detected at 24 and 48 h. The OD value of mock vector-treated 97H cells was significantly lower than that of overexpressed hsa_circ_0002124-treated 97H cells (P<0.01; Figure 4B). Moreover, cell migration and invasion analysis of mock vector-treated and overexpressed hsa_circ_0002124-treated 97H cells revealed that the cell number of cell migration and invasion were significantly higher in overexpressed hsa_circ_0002124 treated-97H cells than in mock vector-treated 97H cells (P<0.05; Figure 4C,F). Furthermore, cell cycle distribution analysis indicated that the cell number of overexpressed hsa_circ_0002124-treated 97L cells in the G1 phase was significantly decreased and that the cell number in the S and G2 phases was significantly increased (P<0.05) compared with mock vector-treated 97L cells, thereby suggesting that overexpressed hsa_circ_0002124 treatments are helpful in promoting the transaction of the cells in the G1 phase to the S and G2 phases (Figure 4D,F). In addition, cell apoptosis of overexpressed hsa_circ_0002124 treated-97H cells decreased the number of cell apoptosis compared with that in the mock vector-treated group (Figure 3E,F) (P<0.01). Our results further suggest that hsa_circ_0002124 facilitates HCC development.
The enrichment GO terms and KEGG
As shown in Figure 5A, the results from GO verified that hsa_circ_0002124 were significantly clustered in Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). The three most significant GO terms in BP were cellular process, biological regulation, and single-organism process, those in CC were cell, cell parts, and cell organelles, and that in MF was binding. In the KEGG analysis, the targets were enriched in the following pathways: Pathways in cancer, PI3K-AKt signaling pathway, HTLV-I infection pathway, and MAPK signaling pathway (Figure 5B).
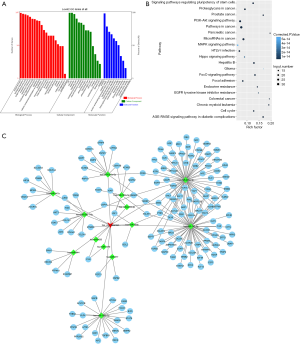
The interaction network
The results in the interaction network showed that hsa_circ_0002124 connected 13 miRNAs, including hsa-miR-944, hsa-miR-620, hsa-miR-577, hsa-miR-4782-3p, hsa-miR-29b-1-5p, hsa-miR-203a-5p, hsa-miR-200a-3p, hsa-miR-190b, hsa-miR-190a-5p, hsa-miR-147a, hsa-miR-141-3p, hsa-miR-135b-5p, and hsa-miR-1270. Through the interaction network construction, 186 target genes were considered to be associated with HCC (Figure 5C), of which the MAPK signaling pathway-related genes were found to be highly frequent.
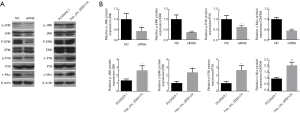
Knockdown and overexpressed hsa_circ_0002124 affected the protein expressions in the MAPK pathway
To further study the protein changes in the MAPK pathway, we performed Western blotting of the knockdown and overexpressed hsa_circ_0002124-treated 97H cells. For knockdown hsa_circ_0002124 treatment, the relative expressions of p-JNK/JNK, p-ERK/ERK, p-P38/P38, and c-Myc in knockdown hsa_circ_0002124-treated 97H cells were significantly lower than those in normal 97H cells (P<0.05) (Figure 6). For overexpressed hsa_circ_0002124 treatments, the relative p-JNK/JNK, p-ERK/ERK, p-P38/P38, and c-Myc expressions in overexpressed hsa_circ_0002124-treated 97H cells were significantly higher than those in mock vector-treated 97H cells (P<0.05) (Figure 6). These results indicate that the hsa_circ_0002124 expression changes in 97H cells are closely associated with the pivotal protein expressions in the MAPK pathway.
Discussion
In the past few decades, although circRNA has been reported as a by-product of splice-mediated splicing errors (9), it had not attracted sufficient attention from researchers (20). However, with the widespread application of high-throughput sequencing technology and bioinformatics, it has been revealed that circRNA is ubiquitous in eukaryotes (21). Increasing evidence has indicated that circRNA could form a closed loop structure by nonlinear reverse splicing, and its expression is the characteristic of tissue-specific, cell-specific, and growth-stage variability (22,23). In cancer, circRNAs can act as a competitive endogenous RNA or miRNA molecular sponge, which could be involved in cancer development (9). Other circRNAs contain a miRNA binding site that could inhibit mRNA degradation by competitively binding to miRNA (24). For example, cir-ITCH can act as a molecular sponge of miR-7, miR-17, and miR-214, inhibiting Wnt/β-Catenin signaling pathway in esophageal squamous cell carcinoma (25). Moreover, ciRS-7 participates in the development and progression of gastric cancer by interacting with miR-7 (26). Furthermore, Hsa_circ_0005386 regulates Notch1 expression by interacting with miR-129-5p in the HCC process, thereby indicating that it can be a potential biomarker for HCC diagnosis (27).
Hsa_circ_0002124 is located at chr15:41667909-41669502, which is the region of intron 9 in NuSAP1. A previous study has shown that NuSAP1 is a microtubule-binding protein that plays an important role in spindle assembly. Its excessive expression leads to the aggregation of microtubules, resulting in a variety of cell arrest and spindle formation inhibition (28). The NuSAP1 mRNA expression levels and protein levels are strictly regulated, which was adapted to the cell cycle process. The NuSAP1 expression reaches its peak in the G2/M period, and the deduction of its expression can lead to stagnation (29). In recent years, the NuSAP1 overexpression has been noted in several tumors, such as pancreatic cancer, melanoma, glioblastoma, childhood acute lymphoblastic leukemia, breast cancer, and acute myeloid leukemia. The NuSAP1 overexpression in tumor is often associated with cancer cell invasiveness (30). For example, Gulzar et al. have reported that recurrence following prostatectomy is significantly associated with transcriptional ascending of NuSAP1. Furthermore, the elevated transcription levels are associated with poor postoperative prognosis in prostate cancer (31). Therefore, NuSAP1 is considered a new biomarker for prostate cancer. Satow et al. have found that the NuSAP1 expression was upregulated in HCC tissues. However, the detailed molecular mechanism remains unclear (32).
In the present study, we revealed that the hsa_circ_0002124 expression was upregulated in HCC cells and clinical samples, which is consistent with the NuSAP1 expression. This evidence is expected to provide a novel clue to the HCC research. Cell functional studies have also suggested that the hsa_circ_0002124 expression is closely associated with cellular proliferation, behavior, cycle, and apoptosis. All these evidences have indicated that hsa_circ_0002124 contains the characteristics that promote HCC development. KEGG and interaction network analyses of hsa_circ_0002124 have indicated that hsa_circ_0002124 could be a molecular sponge of miRNAs, which regulates the key protein expressions in the MAPK pathway. In addition, we assessed the protein expression changes with overexpressed and knockdown hsa_circ_0002124-treated 97H cells. Knockdown hsa_circ_0002124-treated 97H cells showed reduced relative expression of p-JNK/JNK, p-ERK/ERK, p-P38/P38, and c-Myc compared with the normal 97H cells. On the contrary, overexpressed hsa_circ_0002124-treated 97H cells showed increased p-JNK/JNK, p-ERK/ERK, p-P38/P38, and c-Myc expression compared with mock vector-treated 97H cells. The hsa_circ_0002124 expression changes in 97H cells were closely associated with the pivotal protein expressions in the MAPK pathway. HCC is a highly malignant cancer, and elucidating the signaling pathways involved in the development of HCC is essential for the study of new anticancer drugs. Previous studies have shown that ERK, JNK, and p38 and the downstream transcription factors in the MAPK signaling pathway are closely associated with the occurrence and development of liver cancer. For example, the activated ERK signaling pathway plays a role in HCC associated with HBV and HCV infection (33). In JNK knockout mice, the occurrence of DEN-induced HCC is significantly reduced (34). Moreover, the negative regulation of p38 signaling pathway in HCC has been demonstrated in a mouse model with liver-specific p38 knockdown (35). All these evidences support that the core proteins in the MAPK pathway are closely associated with HCC development. In the present study, we speculated that hsa_circ_0002124 is a molecular sponge of miRNA molecular, which regulates the key protein expressions in the MAPK pathway. Therefore, hsa_circ_0002124 is a potential biomarker for HCC diagnosis as well as a novel proliferative factor in HCC. However, the specific regulation mechanism of hsa_circ_0002124 in HCC remains to be further elucidated.
Conclusions
In summary, we confirmed the identification of hsa_circ_0002124 in HCC cells. The hsa_circ_0002124 expression in HCC tissues was significantly higher than in pericarcinomatous tissues. Knockdown and overexpressed hsa_circ_0002124 treatments resulted in changes of cell function, behavior, and apoptosis. Moreover, hsa_circ_0002124 could be a molecular sponge of miRNAs, which regulates the key protein expressions in the MAPK pathway. Our findings are expected to provide detailed understanding regarding the role of hsa_circ_0002124 in HCC.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.01.38). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The present experiment was approved by the Committee on the Ethics of Sun Yat-sen University (approval No. SYSU-20161078). All subjects were completely informed about the study goals and procedure and provided their signed informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 2017;66:1151-64. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Alhasan AA, Izuogu OG, Al-Balool HH, et al. Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood 2016;127:e1-e11. [Crossref] [PubMed]
- Zlotorynski E. Non-coding RNA: Circular RNAs promote transcription. Nat Rev Mol Cell Biol 2015;16:206. [Crossref] [PubMed]
- Szabo L, Morey R, Palpant NJ, et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 2015;16:126. [Crossref] [PubMed]
- Werfel S, Nothjunge S, Schwarzmayr T, et al. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol 2016;98:103-7. [Crossref] [PubMed]
- Yang D, Sun L, Li Z, et al. Noncoding RNAs in Regulation of cancer metabolic reprogramming. Adv Exp Med Biol 2016;927:191-215. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. [Crossref] [PubMed]
- Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta 2016;1859:1245-51. [Crossref] [PubMed]
- Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010;465:1033-8. [Crossref] [PubMed]
- Cesana M, Daley GQ. Deciphering the rules of ceRNA networks. Proc Natl Acad Sci U S A 2013;110:7112-3. [Crossref] [PubMed]
- Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016;7:11215. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- He B, He Y, Shi W, et al. Bioinformatics analysis of gene expression alterations in microRNA-122 knockout mice with hepatocellular carcinoma. Mol Med Rep 2017;15:3681-9. [Crossref] [PubMed]
- Roy S, Hooiveld GJ, Seehawer M, et al. microRNA 193a-5p Regulates Levels of Nucleolar- and Spindle-Associated Protein 1 to Suppress Hepatocarcinogenesis. Gastroenterology 2018;155:1951-1966.e26. [Crossref] [PubMed]
- Papageorgiou I, Court MH. Identification and validation of microRNAs directly regulating the UDP-glucuronosyltransferase 1A subfamily enzymes by a functional genomics approach. Biochem Pharmacol 2017;137:93-106. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Cocquerelle C, Mascrez B, Hétuin D, et al. Mis-splicing yields circular RNA molecules. FASEB J 1993;7:155-60. [Crossref] [PubMed]
- Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014;15:409-39. [Crossref] [PubMed]
- Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet 2013;9:e1003777. [Crossref] [PubMed]
- Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: Identification, biogenesis and function. Biochim Biophys Acta 2016;1859:163-8. [Crossref] [PubMed]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55-66. [Crossref] [PubMed]
- Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget 2015;6:6001-13. [PubMed]
- Pan H, Li T, Jiang Y, et al. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem 2018;119:440-6. [Crossref] [PubMed]
- Fu L, Chen Q, Yao T, et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget 2017;8:43878-88. [PubMed]
- Ribbeck K, Groen AC, Santarella R, et al. NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol Biol Cell 2006;17:2646-60. [Crossref] [PubMed]
- Andersen SS. Spindle assembly and the art of regulating microtubule dynamics by MAPs and Stathmin/Op18. Trends Cell Biol 2000;10:261-7. [Crossref] [PubMed]
- Iyer J, Moghe S, Furukawa M, et al. What's Nu(SAP) in mitosis and cancer? Cell Signal 2011;23:991-8. [Crossref] [PubMed]
- Gulzar ZG, McKenney JK, Brooks JD. Increased expression of NuSAP in recurrent prostate cancer is mediated by E2F1. Oncogene 2013;32:70-7. [Crossref] [PubMed]
- Satow R, Shitashige M, Kanai Y, et al. Combined functional genome survey of therapeutic targets for hepatocellular carcinoma. Clin Cancer Res 2010;16:2518-28. [Crossref] [PubMed]
- Chin R, Earnest-Silveira L, Koeberlein B, et al. Modulation of MAPK pathways and cell cycle by replicating hepatitis B virus: factors contributing to hepatocarcinogenesis. J Hepatol 2007;47:325-37. [Crossref] [PubMed]
- Hui L, Zatloukal K, Scheuch H, et al. Proliferation of human HCC cells and chemically induced mouse liver cancers requires JNK1-dependent p21 downregulation. J Clin Invest 2008;118:3943-53. [Crossref] [PubMed]
- Sakurai T, He G, Matsuzawa A, et al. Hepatocyte necrosis induced by oxidative stress and IL-1 alpha release mediate carcinogen-induced compensatory proliferation and liver tumorigenesis. Cancer Cell 2008;14:156-65. [Crossref] [PubMed]


