
Microarray-based differential expression profiling of long noncoding RNAs and messenger RNAs in formalin-fixed paraffin-embedded human papillary thyroid carcinoma samples
Introduction
In recent decades, the incidence of thyroid cancer has increased in most regions of the world (1,2). Among different forms of thyroid cancer, papillary thyroid carcinoma (PTC) is the most common form, accounting for 85–95% of thyroid tumors (3,4). Several population-based studies have revealed that while rates of thyroid cancer have increased, small tumors are now more likely to be detected and there have been no significant changes in mortality associated with this disease (5,6). A family history of thyroid cancer, exposure to radiation, and abnormal intake of iodine are all PTC risk factors (7). Recent studies have indicated that genomic instability, alterations in epigenetic regulation, and subsequent inappropriate gene expression have key roles in the regulation of PTC (8,9). However, the mechanisms underlying PTC pathogenesis have not been fully elucidated to date. In clinical contexts, nodular goiters are known to be capable of developing into poorly differentiated thyroid carcinoma (PDC), but the genomic mechanisms governing this transformation remain uncertain. A thorough investigation of the mechanisms underlying this transformation could thus offer new clinical insights into the pathogenesis or treatment of PDC.
The majority of genes are regulated through complex epigenetic mechanisms, with a diverse array of noncoding RNAs (ncRNAs) participating in these regulatory processes (10,11). In the context of PTC, the functions of specific microRNAs have been relatively well elucidated, but alterations in PTC-associated long noncoding RNAs (lncRNAs) have also been found to lead to the altered regulation of key genes, and these events are less well understood (12,13). Studies have shown that aberrant gene expression and the tumorigenesis can be caused by dysregulated expression of lncRNAs. In addition, in many kinds of cancers, changes in lncRNA expression can accelerate the invasion and metastatic progression of the disease (14). At present, the study on the role of lncRNAs in PTC progression is in its early stages, and further studies are needed to explore the lncRNAs associated with PTC and their mechanisms of action.
Intracellular mammalian metallothioneins (MTs) are low molecular weight proteins (~6 kDa) initially isolated from horse renal cortex tissue in 1957 by Margoshes and Valee (15,16). In humans, of the 17 different genes encoding MTs, 13, 2, 1, and 1 gene each encode MT-1, MT-2, MT-3, and MT-4, respectively (17-19). MT is primarily located in the cytoplasm of adult tissues, while in fetal/early neonatal tissues it is also found in the nuclei of cells (20). MT located in the nucleus has been observed during cell proliferation and differentiation (21). Expression of MT family genes and altered localization of MT in cells during development suggest that MT may be a proto-oncogene. The carcinogenic potential of MT has been evaluated in workshops and symposiums (22), and several reports have studied MT expression in human tumors (23).
In the current study, the expression of lncRNAs and mRNAs were detected in seven PTC and seven nodular goiter samples via a microarray analysis, and differentially expressed lncRNAs and mRNAs associated with PTC were identified. In addition, we demonstrated that genes associated with these lncRNAs and mRNAs were known or suspected risk factors for tumorigenesis. These differentially expressed lncRNAs and mRNAs may thus be novel molecular biomarkers useful for the diagnosis and/or treatment of PTC.
Methods
Patient samples and hematoxylin & eosin (H&E) staining
Seven patients with a follicular variant of papillary thyroid carcinoma (FVPTC) (2 males and 5 females, mean age: 39.43±13.06 y) and seven patients with nodular goiter, none of whom had undergone radiation, chemotherapy, or immunotherapy treatments, were recruited from Beijing Haidian Hospital Beijing Haidian Section of Peking University Third Hospital in China between January 2011 and January 2013. The diagnosis of PTC was made by at least two experienced oncologists. Sample collection was conducted according to the following criteria: the minimum tumor diameter was greater than 2 cm. The Formapure nucleic acid extraction kit (Agencourt Biosciences, Beverly, MA, USA) was then used to extract and purify the RNA from formalin-fixed, paraffin-embedded (FFPE) tissue sections prepared from these tumor samples. TRIzol (Invitrogen, Carlsbad, CA, USA) was used to extract RNA from frozen tissues and the RNeasy Protect kit (Qiagen, Valencia, CA, USA) was used to purify RNA from these samples. To remove contaminating DNA from RNA samples, DNase I treatment (Ambion, Austin, TX, USA) was additionally used. RNA concentrations were determined using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Rockland, DE, USA), and electropherograms were used to evaluate the integrity of RNA, with the RNA 6000 PicoAssay for the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) being chosen to determine the RNA integrity number, RIN42 (a correlative measure of mRNA intactness). Hematoxylin and eosin (H&E) staining was conducted as previously described. Briefly, FFPE tissue samples were sectioned at 3–5 µm thickness and stained with stained with hematoxylin (Sigma H3136) for 10 min and with eosin (Sigma E4382) for 1 min to better visualize diagnostic areas.
Microarray and computational analysis
Sample labeling and array hybridization were performed according to provided protocols. Briefly, after the rRNA was removed, the mRNA-ONLY™ Eukaryotic mRNA Isolation kit (Epicentre, Madison, WI, USA) was used to purify mRNA from total RNA. Subsequently, each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3' bias utilizing a random priming method (Flash RNA Labeling kit; Arraystar, Rockville, MD, USA). The RNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA) was then used to purify the labeled cRNAs. A NanoDrop ND-1000 was used to measure concentrations of the labeled cRNAs and their specific activity (pmol Cy3/µg cRNA). Labeled cRNAs were then hybridized onto the Affymetrix GeneChip® Human Transcriptome Array 2.0. lncRNAs were carefully constructed using the following public transcriptome databases: RefSeq (NCBI Reference Sequence Database; www.ncbi.nlm.nih.gov/refseq/); NCBI unigene (https://www.ncbi.nlm.nih.gov/unigene); Ensembl (http://ensemblgenomes.org/); UCSC (genome.ucsc.edu/index.html); LNCRNA-DB (http://www.lncrnadb.org/); as well as landmark publications (24). To identify each transcript accurately, a specific exon or splice junction probe was applied, and subsequently the hybridized arrays were washed and fixed. To analyze acquired array images, the Affymetrix DNA Microarray Scanner and Feature Extraction software was used to scan the fixed hybridized arrays. Subsequently, a variation of the reads/Kb/Million method and Z-score analysis were performed to normalize the raw data. Differences between microarray data for the two groups were compared via Student’s t-test. Significance of these microarray results was analyzed based on fold change (FC), and P values were corrected based on a false discovery rate (FDR)]. The threshold value used to define differentially expressed lncRNAs and mRNAs was a FC of ≥2.0 or ≤0.5, with P<0.05 as the threshold of statistically significance. SPSS v13.0 (SPSS, Inc., Chicago, IL, USA) was used for all statistical analyses.
Gene ontology (GO) and pathway analysis
To determine the functions of differentially expressed mRNAs and their associated pathways, we used the local FunNet software to analyze the GO term and pathway enrichment based on the available Gene Ontology platform (www.geneontology.org), which provides three structured networks of defined items: biological processes, cellular components, and molecular functions. In this analysis, the P value denotes the significance of enrichment of a given GO term in the differentially expressed mRNA list (cut-off: P<0.05). To highlight important pathways, pathways enriched in differentially expressed mRNAs were analyzed based on the latest kyoto encyclopedia of genes and genomes pathway annotations (www.genome.jp/kegg/). For the KEGG analysis, P values denote the significance of the pathway associated with the latest data available in the KEGG database (cut-off, P<0.05).
Hierarchical clustering analysis and co-expression analysis
Hierarchical clustering was performed to analyze the significantly differentially expressed lncRNA and mRNA datasets as previously described by Eisen et al. (25). Briefly, a log2 transformation was performed for the relative ratios of significantly aberrantly expressed lncRNAs and mRNAs, and then similarity was defined based on the Euclidean distance similarity metric, with the complete linkage clustering method chosen to assemble hierarchical clusters. The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database of physical and functional interactions (26) was then used to analyze protein-protein interactions (PPIs), and the Cytoscape software (version 3.0.0) was used to represent the resultant PPI network (27).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Differential expression of select mRNAs identified in the initial microarray was verified by RT-qPCR. In total, three mRNAs (down-regulated: MT1G, MT1F, and MT1H) were chosen for a q-PCR reaction in twenty PTC and nodular goiter samples. TRIzol was used to extract total RNA from forty FFPE specimens, and this RNA was reverse transcribed into cDNA using the PrimeScript RT reagent kit (Invitrogen), according to provided instructions. Samples were also treated with DNase (1 µL; 1U/µL) to remove contaminating DNA. qPCR reactions were then carried out in a total of 50 µl volume per well. qPCR settings were as follows: 95 °C for 2 min; 95 °C for 15 sec, 57 °C for 15 sec, and 72 °C for 1 min for a total 40 cycles. The Taqman probes used for the three mRNAs were as follows: ACTB: 5'-CCTTTGCCGATCCGCCGCCCGTCCA-3'; MT1G: 5'-GAAGTGCAGCTGCTGCGCCTGATGT-3' and 5'-GCTCCTGTGCCGCTGCAGGTGTCTC-3'; MT1F: 5'-TTATACCACCTTGACCCATTTGCTA-3'; and MT1H: 5'-TACAACTCCGACTCATTTGCTACAT-3'. Data were collected using the ABI analytical thermal cycler.
Results
Differential expression profiles of lncRNAs and mRNAs in PTC
Figure 1 shows the typical pathological characteristics of patients with PTC. PTC was identified in patient samples via H&E staining, and the ratio of cancer cells to total cells in individual tissue sections was at least 80%. PTC tissue and nodular goiter tissues in each patient were collected, fixed, and paraffin embedded. The human PTC and nodular goiter tissue samples were then analyzed by microarray to explore the differential expression of lncRNAs and mRNAs between these two sample types, with goiter samples serving as the experimental control tissue. The normalized expression values from the nodular goiter tissue samples were used to calculate the FCs and P values for each gene. Relative to the nodular goiter tissue samples, there were significantly differences in the expression of 94 lncRNAs and 89 mRNAs in PTC samples (FC ≥2.0; P<0.01). Among the 94 differently expressed lncRNAs, 15 were up-regulated and 79 were down-regulated in PTC samples. Among the 89 abnormally expressed mRNAs, 12 were up-regulated and 77 were down-regulated (Figure 2, http://fp.amegroups.cn/cms/tcr.2019.02.12-1.pdf). Based on the differential expression levels of the identified lncRNAs and mRNAs, both were arranged into groups via a hierarchical clustering analysis (Figure 3). The expression modes of lncRNA and mRNAs were further explored using a dendrogram (FC ≥2.0; P<0.01; Figure 2). n340297 (FC =3.7) was the most down-regulated lncRNA, while n332905 (FC =2.7) was the most up-regulated lncRNA. MT1G (FC =10.9) was the most down-regulated mRNA, while SLC34A2 (FC =4.3) was the most up-regulated mRNA. In total we have summarized 40 significantly differential expressed items, including 20 significantly up-regulated expressed items and 20 significantly down-regulated expressed items (Table 1). Notably, multiple members of the MT gene family were all down-regulated in PTC samples as compared with nodular goiter tissue samples (Table 2).
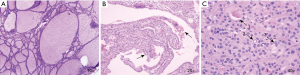
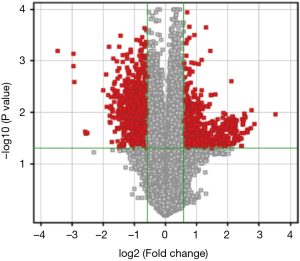
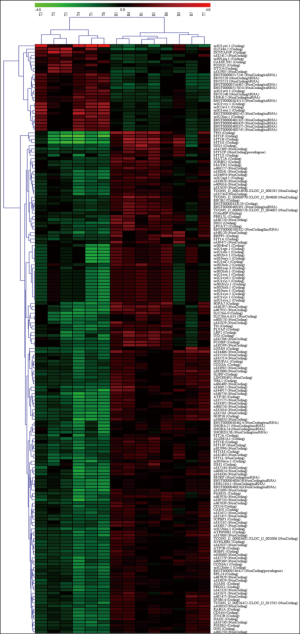
Table 1
| Primary ID | Fold changes | Regulation | P value | Locus type |
|---|---|---|---|---|
| MT1G | 10.94924 | Down | 6.45E-04 | Coding |
| MT1H | 7.715949 | Down | 7.24E-04 | Coding |
| MT1F | 7.657472 | Down | 0.001268 | Coding |
| TPO | 7.596827 | Down | 0.002575 | Coding |
| SLC2644 | 4.001563 | Down | 0.003759 | Coding |
| IYD | 3.731338 | Down | 0.001858 | Coding |
| n340297 | 3.698557 | Down | 0.007797 | Non-coding |
| n407932 | 3.568332 | Down | 0.004669 | Non-coding |
| n342369 | 3.523873 | Down | 5.32E-04 | Non-coding |
| n336033 | 3.39254 | Down | 0.008311 | Non-coding |
| n334406 | 3.364263 | Down | 0.007475 | Non-coding |
| LRP2 | 3.35106 | Down | 0.00515 | Coding |
| HSPA5 | 3.230271 | Down | 0.009589 | Coding |
| n344917 | 3.22871 | Down | 0.003499 | Non-coding |
| DIO1 | 3.213385 | Down | 0.008882 | Coding |
| n339733 | 3.177889 | Down | 9.03E-04 | Non-coding |
| n336012 | 3.147624 | Down | 0.003718 | Non-coding |
| n340776 | 3.144596 | Down | 0.006244 | Non-coding |
| ATP5H | 3.117614 | Down | 0.002836 | Coding |
| nN335616 | 3.087649 | Down | 0.004551 | Non-coding |
| uc021yst.1 | 6.794604 | Up | 0.009216 | Coding |
| SLC34A2 | 4.349818 | Up | 0.002504 | Coding |
| ENST00000408542 | 3.050107 | Up | 0.004546 | Non-coding/miRNA |
| uc021vey.1 | 2.737648 | Up | 0.004626 | Coding |
| uc022avi.1 | 2.737648 | Up | 0.004626 | Coding |
| n332905 | 2.712229 | Up | 6.49E-04 | Non-coding |
| DCSTAMP | 2.650783 | Up | 0.006883 | Coding |
| uc021wsr.1 | 2.580299 | Up | 0.007814 | Coding |
| ENST00000517241 | 2.471056 | Up | 0.007653 | Non-coding/snRNA |
| n332412 | 2.446805 | Up | 2.29E-04 | Non-coding |
| ENST00000408429 | 2.311055 | Up | 0.007908 | Non-coding/miRNA |
| ENST00000401335 | 2.259779 | Up | 0.008486 | Non-coding/miRNA |
| ENST00000517054 | 2.25374 | Up | 0.008552 | Non-coding/snRNA |
| MIR412 | 2.230374 | Up | 0.003681 | Non-coding/miRNA |
| ENST00000516485 | 2.22826 | Up | 0.006558 | Non-coding/snRNA |
| CAMK2N1 | 2.21727 | Up | 9.50E-04 | Coding |
| DTX4 | 2.213393 | Up | 0.004628 | Coding |
| ENST00000408375 | 2.155374 | Up | 0.008106 | Non-coding/miRNA |
| uc022bin.1 | 2.138922 | Up | 0.006837 | Coding |
| ENST00000408745 | 2.110623 | Up | 0.009567 | Non-coding/miRNA |
Table 2
| Gene symbol | Fold changes | Regulation | P value | Locus type |
|---|---|---|---|---|
| MT1G | 10.94924 | Down | 6.45E-04 | Coding |
| MT1H | 7.715949 | Down | 7.24E-04 | Coding |
| MT1F | 7.657472 | Down | 0.001268 | Coding |
| MT1L | 2.810005 | Down | 0.001391 | Non-coding/non-coding RNA |
| MT1JP | 2.789976 | Down | 5.35E-04 | Non-coding/non-coding RNA |
| MT1X | 2.528777 | Down | 0.006529 | Coding |
| MT1M | 2.348179 | Down | 7.47E-04 | Coding |
| MT1CP | 2.328058 | Down | 9.09E-04 | Non-coding/pseudogene |
| MT1E | 2.255994 | Down | 0.002713 | Coding |
| MT1A | 2.147448 | Down | 6.96E-04 | Coding |
| MT1B | 1.49531 | Down | 0.001379 | Coding |
PTC, papillary thyroid carcinoma.
GO and pathway analysis
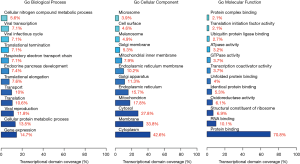
GO terms were further assigned to the significantly differentially expressed genes based on their sequence similarities with known proteins in the UniProt database. Genes were annotated both with GO terms as well as with the InterPro and Pfam domains contained therein. GO annotation and enrichment analysis of twelve differentially expressed sequences including MT1G, MT1H, MT1F, TPO, uc021yst, SLC34A2, SLC26A4, IYD, n340297, n407932, n342369 and n336033 was conducted using the local FunNet software (28), which corrected for gene length bias. GO terms with corrected P values <0.05 were considered to be significantly enriched among these differential expressed genes (http://fp.amegroups.cn/cms/tcr.2019.02.12-2.pdf, http://fp.amegroups.cn/cms/tcr.2019.02.12-3.pdf, http://fp.amegroups.cn/cms/tcr.2019.02.12-4.pdf). The bar charts of GO biological processes, GO cellular components, and GO molecular functions distributions are shown in Figure 4. With respect to molecular functions, 96 GO terms were assigned among which the top three significantly enriched terms were protein binding (GO: 0005515, P value =6.14E-22), RNA binding (GO: 0003723, P value =4.35E-05), and structural constituent of ribosome (GO: 0003735, P value =3.76E-12). With respect to cellular components, 70 GO terms were assigned of which the most significantly enriched terms were cytoplasm (GO: 0005737, P value =5.65E-03), membrane (GO: 0016020, P value =1.86E-03), and cytosol (GO: 0005829, P value =8.28E-13). With respect to biological processes, 237 GO terms were assigned among which gene expression (GO: 0010467, P value =7.93E-16) and metabolic process of cellular protein (GO: 0044267, P value =1.86E-19) were the top two significantly over-represented terms (Figure 4).
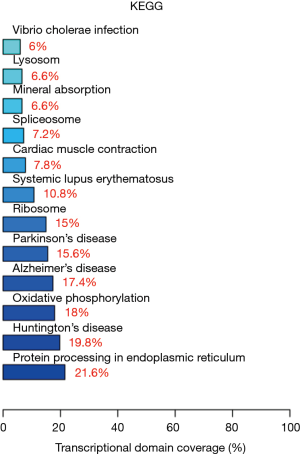
In vivo, biological functions arise from complex and interdependent regulation of multiple different signaling pathways. Pathways enrichment analysis can offer some clues with respect to the biochemical and signal transduction pathways in which differentially expressed genes may participate. The KEGG database can thus be used to understand the high-level functions of a given biological system, such as a cell, an organism, or an ecosystem using large-scale molecular datasets generated by genome sequencing and other high-through put experimental technologies (http://www.genome.jp/kegg/). The local FunNet software was used to test the statistical enrichment of differentially expressed genes in particular KEGG pathways. In this study, 11 differentially expressed genes were found to be involved in 18 pathways (http://fp.amegroups.cn/cms/tcr.2019.02.12-5.pdf). Figure 5 shows the results of the pathway enrichment analysis, clearly revealing that protein processing in endoplasmic reticulum (04141, P value =4.3E-14) was the top enriched item. Eight differentially expressed genes identified in our study participate in this pathway. In addition, oxidative phosphorylation, Huntington’s disease, and Alzheimer’s were also significant enriched in this study.
RT-qPCR and co-expression analysis
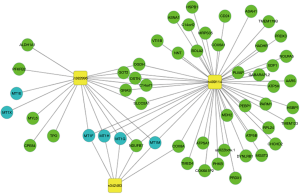
To verify the microarray data, three significantly down-regulated mRNAs (MT1G, MT1F, and MT1H) mRNAs were randomly selected for an RT-qPCR analysis in 20 PTC and 20 nodular goiter tissue samples. The results of this analysis were consistent with the microarray data (Figure 6). Moreover, to identify the regulatory networks dictating relationships between the differentially expressed lncRNAs and mRNAs and their potential substrates a co-expression analysis was performed using the STRING database. To improve the accuracy of this analysis, we set the confidence level (score) to a high value (0.900). Then, using the Cytoscape software, the network of all lncRNAs and mRNAs with their potential substrates was extracted from the whole interaction network and reconstructed. Based on Pearson correlation values (Pearson correlation >0.90, P value <0.01), we further screen out 57 lncRNAs and mRNAs which participated in the regulatory network. Of note, n382996, n342483 and n409114 were closely associated with MT1G, MT1H and MT1F. Based on these results, we speculate that these lncRNA may participate in the regulation of the MT gene family during PTC development or progression (Figure 7).

Discussion
PTC is a common endocrine cancer that occurs more often in women (29). The incidence of PTC has been increasing worldwide in recent decades. PTC tumorigenesis is an intricate process with many differentially expressed genes contributing to the development of disease. Many studies have extensively explored the molecular mechanism of PTC. However, the exact pathogenesis of this disease is not fully understood. In clinical settings, nodular goiter tissues can develop into PDC, but the genomic mechanisms underlying this transformation are poorly understood. Studies of gene expression in thyroid tumors offer insights into the molecular differences among different thyroid disease types. In a valuable study performed by The Cancer Genome Atlas Research Network, the genomic landscape of PTC has been comprehensively described, revealing that compared with other solid tumor types, PTC has a lower frequency of somatic mutations. CHEK2, ATM, TERT, miR-21, and miR-146b all contribute to the clinical classification and progression of this disease. Eszlinger et al. have described the differences in gene expression between the functional and nonfunctional benign thyroid tumors (30). In a separate study performed by Huang et al. (31) using high density DNA microarrays to analyze PTC samples, certain markers identified in this study were validated as PTC markers (32). Recent evidence strongly suggests that lncRNAs play a vital role in regulating gene expression. Although lncRNAs were initially considered to be a form of transcriptional noise, studies have more recently revealed their significant role in cellular development and various diseases (33,34), including a role for lnc-IL7R in inflammation (35), and for HOTAIR and Gas5 in breast tumors and metastases (36,37). However, in the pathogenesis and development of PTC, the role of lncRNAs is not well understood. Our present study revealed differentially expression profiles for both lncRNAs and mRNAs in PTC tumor samples, comparing the expression level of these transcripts with those in nodular goiter tissue. In total, 94 lncRNAs and 89 mRNAs were significantly differentially expressed in PTC samples. Among the 94 lncRNAs, 15 were up-regulated and 79 were down-regulated. Among the 89 mRNAs, 12 were up-regulated and 77 were down-regulated (Figure 2 and http://tcr.amegroups.com/public/system/tcr/supp-tcr.2019.02.12-1.pdf). A dendrogram revealed the lncRNA and mRNAs expression patterns among samples (FC ≥2.0; P<0.01; Figure 2). n340297 (FC =3.7) was the most down-regulated lncRNA, while n332905 (FC =2.7) was the most up-regulated lncRNA. Additionally, MT1G (FC =10.9) was the most down-regulated mRNA and SLC34A2 (FC =4.3) was the most up-regulated mRNA.
The results of the present study revealed an altered lncRNA and mRNA expression profile in PTC. In our study, we found that the genes related to our observed differentially expressed lncRNAs and mRNAs were associated with many kinds of cancer. These genes included MT1s, which were significantly down-regulated in PTC and in several other types of cancer. A study found that among 34 human thyroid tumors 31 showed either nuclear and/or cytoplasmic localization of MT, while less than 20% of the normal thyroid gland tissue exhibited diffuse MT distribution (38). Compared to normal thyroid, down-regulated MT-I+II genes have been found both in papillary and follicular thyroid carcinomas by microarray analysis (39). When MT-1s expression was restored to malignant cells via cDNA transfection, cells growth rates and tumorigenicity were suppressed in vivo (39), indicating a suppressive role for MT-1s in thyroid carcinogenesis. This was consistent with a previous study which found that MT-1s was down-regulated in PTC samples relative to normal thyroid tissue owing to the hypermethylation of the MT-1s gene promoter. Furthermore, a negative correlation was detected between MT-1s and poor thyroid cancer prognosis in patients with other poor prognostic factors (40). MT-1s is also vital to many cellular processes, regulating the homeostasis of zinc ions and thus potentially regulating the activity of multiple zinc-dependent transcription factors and enzymes, such as metalloproteinase (41,42). There is abundant evidence that MT-1s plays a role in cell proliferation—MT-1s can be observed in the nuclei of cells in certain stages of the cell cycle, whereas MT-1s is primarily nuclear in quiescent cells (43). Biochemical studies have found that the concentrations of MT-1s are highest in the cytoplasm in the G1 phase, while MT-1s translocates into the nucleus during the G1/S phase (21). Furthermore, in the S and G2 phases the concentration of MT-1s was highest, supporting a role for this protein in cell division (44). Cytoplasmic-nuclear expression of MT-1s has been noted in regenerating kidney (45) and liver tissue (45,46), as well as in proliferating basal and parabasal cells of stratified epithelia (47). The role of MT-1s in the activation of cell proliferation is also supported by studies that found a positive correlation between MT-1s and the expression Ki-67 antigen in various tumors, including in breast cancer (48), squamous and basal cell skin cancers (49), adrenocortical cancers (ACC) (50), non-small cell lung cancers (NSCLC) (51), gastrointestinal stromal tumors (GISTs) (52), and sarcomas (53). Many studies have described the important role of MT-1s in protecting DNA against free radical damage (54) induced by radiation (55) or chemotherapeutic agents, such as cisplatin, etoposide, melfalan, bleomycin, or irinotecan administered during the treatment of breast, ovarian, gastric, or prostate cancer (56,57). Furthermore, studies have also highlighted the stimulatory role of MT-1s during angiogenesis and collaterogenesis under stress conditions such as hypoxia (58). In hypoxic conditions, MT-1s expression increased, and it was able to stabilize the expression of hypoxia-inducible factor-1a (HIF-1a) (59). MT-1s can also induce proangiogenic factors such as fibroblast growth factor (FGF), transforming growth factor b (TGF-b), and vascular endothelial growth factor A (VEGF-A) (60). In spite of these cytoprotective activities, the over-expression of MT-1s is linked with an unfavorable prognosis in several cancers including NSCLC (61), intrahepatic cholangiocarcinoma (ICC) (62), renal cancer (63), ovarian cancer (64), and melanoma (65). However, the prognostic value of MT-1s seems to be limited in some tumors, such as thyroid or laryngeal cancer (66). Some isoforms of MT-1s have been shown to be cancer suppressors. The MT-1G isoform has been found to be a potent oncosuppressor in papillary thyroid (39) and hepatocellular cancers (67), and MT-1F may play a suppressive role in colon cancer (68).
GO annotation and enrichment analysis of twelve differentially expressed transcripts including MT1G, MT1H, MT1F, TPO, uc021yst, SLC34A2, SLC26A4, IYD, n340297, n407932, n342369, and n336033 was implemented using the local FunNet software (28), with results shown in Figure 4. With respect to molecular functions, 96 GO terms were assigned of which the three most significantly enriched terms were protein binding, RNA binding, and structural constituent of ribosome. With respect to cellular components, 70 GO terms were assigned of which cytoplasm, membrane, and cytosol were the most significantly enriched. With respect to biological processes, 237 GO terms were assigned of which gene expression and cellular protein metabolic process were the top two significantly over-represented terms. A KEGG analysis revealed that protein processing in the endoplasmic reticulum was the most enriched pathway among differentially expressed transcripts, with eight of these transcripts participating in this pathway. In addition, oxidative phosphorylation, Huntington’s disease, and Alzheimer’s disease was also significant enriched in this pathway analysis.
In conclusion, our study characterized the significantly altered expression profiles of lncRNAs and mRNAs in PTC. Abnormal regulation of these altered lncRNAs and mRNA may play vital role in the development, progression, and metastasis of this disease. Further research will be needed to explore whether these alterations are linked with genotype, familial cancer risk, inflammatory states, or a combination of these cancer risk factors. In addition, further study of the molecular mechanisms in which these mRNAs and lncRNAs participate may yield further insights into PTC pathogenesis.
Acknowledgments
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.02.12). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethical Committee of PLA General Hospital and Beijing Haidian Hospital Beijing Haidian Section of Peking University Third Hospital (C2015-013-01). All procedures were conducted in accordance with the relevant guidelines and regulations. All patients were made aware of the goals of this study, and provided written informed consent. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- La Vecchia C, Malvezzi M, Bosetti C, et al. Thyroid cancer mortality and incidence: a global overview. Int J Cancer 2015;136:2187-95. [Crossref] [PubMed]
- Chan JK. The wonderful colors of the hematoxylin-eosin stain in diagnostic surgical pathology. Int J Surg Pathol 2014;22:12-32. [Crossref] [PubMed]
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014;64:9-29. [Crossref] [PubMed]
- Yang L, Zheng R, Wang N, et al. Analysis of incidence and mortality of thyroid cancer in China, 2010). Zhonghua Yu Fang Yi Xue Za Zhi 2014;48:663-8. [PubMed]
- Hodgson NC, Button J, Solorzano CC. Thyroid cancer: is the incidence still increasing? Ann Surg Oncol 2004;11:1093-7. [Crossref] [PubMed]
- Zhu C, Zheng T, Kilfoy BA, et al. A birth cohort analysis of the incidence of papillary thyroid cancer in the United States, 1973-2004. Thyroid 2009;19:1061-6. [Crossref] [PubMed]
- Leux C, Truong T, Petit C, et al. Family history of malignant and benign thyroid diseases and risk of thyroid cancer: a population-based case-control study in New Caledonia. Cancer Causes Control 2012;23:745. [Crossref] [PubMed]
- Santos JC, Bastos AU, Cerutti JM, et al. Correlation of MLH1 and MGMT expression and promoter methylation with genomic instability in patients with thyroid carcinoma. BMC Cancer 2013;13:79. [Crossref] [PubMed]
- Neta G, Brenner AV, Sturgis EM, et al. Common genetic variants related to genomic integrity and risk of papillary thyroid cancer. Carcinogenesis 2011;32:1231-7. [Crossref] [PubMed]
- Taft RJ, Pang KC, Mercer TR, et al. Non-coding RNAs: regulators of disease. J Pathol 2010;220:126-39. [Crossref] [PubMed]
- Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature 2012;489:101-8. [Crossref] [PubMed]
- Jendrzejewski J, He H, Radomska HS, et al. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci U S A 2012;109:8646-51. [Crossref] [PubMed]
- Guo Q, Zhao Y, Chen J, et al. BRAF-activated long non-coding RNA contributes to colorectal cancer migration by inducing epithelial-mesenchymal transition. Oncol Lett 2014;8:869-75. [Crossref] [PubMed]
- Zhu M, Chen Q, Liu X, et al. lncRNA H19/miR‐675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J 2014;281:3766-75. [Crossref] [PubMed]
- Sutherland DE, Stillman MJ. The "magic numbers" of metallothionein. Metallomics 2011;3:444-63. [Crossref] [PubMed]
- Theocharis S, Klijanienko J, Giaginis C, et al. Metallothionein expression in mobile tongue squamous cell carcinoma: associations with clinicopathological parameters and patient survival. Histopathology 2011;59:514-25. [Crossref] [PubMed]
- Mididoddi S, McGuirt JP, Sens MA, et al. Isoform-specific expression of metallothionein mRNA in the developing and adult human kidney. Toxicol Lett 1996;85:17-27. [Crossref] [PubMed]
- Quaife CJ, Findley SD, Erickson JC, et al. Induction of a new metallothionein isoform (MT-IV) occurs during differentiation of stratified squamous epithelia. Biochemistry 1994;33:7250-9. [Crossref] [PubMed]
- Palmiter RD, Findley SD, Whitmore TE, et al. MT-III, a brain-specific member of the metallothionein gene family. Proc Natl Acad Sci U S A 1992;89:6333-7. [Crossref] [PubMed]
- Chan HM, Cherian MG. Ontogenic changes in hepatic metallothionein isoforms in prenatal and newborn rats. Biochem Cell Biol 1993;71:133-40. [Crossref] [PubMed]
- Cherian MG, Apostolova MD. Nuclear localization of metallothionein during cell proliferation and differentiation. Cell Mol Biol (Noisy-le-grand) 2000;46:347-56. [PubMed]
- Cherian MG, Howell SB, Imura N, et al. Role of metallothionein in carcinogenesis. Toxicol Appl Pharmacol 1994;126:1-5. [Crossref] [PubMed]
- Cherian MG. The significance of the nuclear and cytoplasmic localization of metallothionein in human liver and tumor cells. Environ Health Perspect 1994;102:131-5. [PubMed]
- Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Gene Dev 2011;25:1915-27. [Crossref] [PubMed]
- Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 1998;95:14863-8. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 2011;39:D561-8. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Prifti E, Zucker JD, Clement K, et al. FunNet: an integrative tool for exploring transcriptional interactions. Bioinformatics 2008;24:2636-8. [Crossref] [PubMed]
- Kweon SS, Shin MH, Chung IJ, et al. Thyroid cancer is the most common cancer in women, based on the data from population-based cancer registries, South Korea. Jpn J Clin Oncol 2013;43:1039-46. [Crossref] [PubMed]
- Eszlinger M, Krohn K, Paschke R. Complementary DNA expression array analysis suggests a lower expression of signal transduction proteins and receptors in cold and hot thyroid nodules. J Clin Endocrinol Metab 2001;86:4834-42. [Crossref] [PubMed]
- Huang Y, Prasad M, Lemon WJ, et al. Gene expression in papillary thyroid carcinoma reveals highly consistent profiles. Proc Natl Acad Sci U S A 2001;98:15044-9. [Crossref] [PubMed]
- Prasad ML, Pellegata NS, Kloos RT, et al. CITED1 protein expression suggests Papillary Thyroid Carcinoma in high throughput tissue microarray-based study. Thyroid 2004;14:169-75. [Crossref] [PubMed]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Gene Dev 2009;23:1494-504. [Crossref] [PubMed]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009;10:155-9. [Crossref] [PubMed]
- Cui H, Xie N, Tan Z, et al. The human long noncoding RNA lnc‐IL7R regulates the inflammatory response. Eur J Immunol 2014;44:2085-95. [Crossref] [PubMed]
- Bhan A, Hussain I, Ansari KI, et al. Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J. Steroid Biochem. Mol Biol 2014;141:160-70. [Crossref] [PubMed]
- Mourtada-Maarabouni M, Pickard M, Hedge V, et al. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 2009;28:195-208. [Crossref] [PubMed]
- Nartey N, Cherian MG, Banerjee D. Immunohistochemical localization of metallothionein in human thyroid tumors. Am J Pathol 1987;129:177-82. [PubMed]
- Ferrario C, Lavagni P, Gariboldi M, et al. Metallothionein 1G acts as an oncosupressor in papillary thyroid carcinoma. Lab Invest 2008;88:474-81. [Crossref] [PubMed]
- Huang 1, de la Chapelle A, Pellegata NS. Hypermethylation, but not LOH, is associated with the low expression of MT1G and CRABP1 in papillary thyroid carcinoma. Int J Cancer 2003;104:735-44. [Crossref] [PubMed]
- Ostrakhovitch EA, Olsson PE, Hofsten JV, et al. P53 mediated regulation of metallothionein transcription in breast cancer cells. J Cell Biochem 2007;102:1571-83. [Crossref] [PubMed]
- Kim HG, Jin YK, Han EH, et al. Metallothionein-2A overexpression increases the expression of matrix metalloproteinase-9 and invasion of breast cancer cells. Febs Letters 2011;585:421-8. [Crossref] [PubMed]
- Apostolova MD, Ivanova IA, Cherian MG. Signal transduction pathways, and nuclear translocation of zinc and metallothionein during differentiation of myoblasts. Biochem Cell Biol 2000;78:27-37. [Crossref] [PubMed]
- Levadoux-Martin M, Hesketh J, Jh Wallace H. Influence of metallothionein-1 localization on its function. Biochem J 2001;355:473-9. [Crossref] [PubMed]
- Zalups RK, Fraser J, Koropatnick J. Enhanced transcription of metallothionein genes in rat kidney: effect of uninephrectomy and compensatory renal growth. Am J Physiol 1995;268:F643-50. [PubMed]
- Cherian MG, Kang YJ. Metallothionein and liver cell regeneration. Exp Biol Med (Maywood) 2006;231:138-44. [Crossref] [PubMed]
- Zamirska A, Matusiak Ł, Dziegiel P, et al. Expression of metallothioneins in cutaneous squamous cell carcinoma and actinic keratosis. Pathol Oncol Res 2012;18:849-55. [Crossref] [PubMed]
- Wojnar A, Pula B, Piotrowska A, et al. Correlation of intensity of MT-I/II expression with Ki-67 and MCM-2 proteins in invasive ductal breast carcinoma. Anticancer Res 2011;31:3027-33. [PubMed]
- Bieniek A, Pula B, Piotrowska A, et al. Expression of metallothionein I/II and Ki-67 antigen in various histological types of basal cell carcinoma. Folia Histochem Cytobiol 2012;50:352-7. [Crossref] [PubMed]
- Szajerka A, Dziegiel P, Szajerka T, et al. Immunohistochemical evaluation of metallothionein, Mcm-2 and Ki-67 antigen expression in tumors of the adrenal cortex. Anticancer Res 2008;28:2959-65. [PubMed]
- Werynska B, Pula B, Muszczynska-Bernhard B, et al. Correlation between expression of metallothionein and expression of Ki-67 and MCM-2 proliferation markers in non-small cell lung cancer. Anticancer Res 2011;31:2833-9. [PubMed]
- Puła B, Strutyńskakarpińska M, Markowskawoyciechowska A, et al. Expression of metallothionein and Ki-67 antigen in GISTs of different grade of malignancy. Adv Clin Exp Med 2013;22:513-8. [PubMed]
- Dziegiel P, Salwa-Zurawska W, Zurawski J, et al. Prognostic significance of augmented metallothionein (MT) expression correlated with Ki-67 antigen expression in selected soft tissue sarcomas. Histol. Histopathol 2005;20:83-9. [PubMed]
- Ruttkay-Nedecky B, Nejdl L, Gumulec J, et al. The role of metallothionein in oxidative stress. Int J Mol Sci 2013;14:6044-66. [Crossref] [PubMed]
- Cai L, Cherian MG. Zinc-metallothionein protects from DNA damage induced by radiation better than glutathione and copper- or cadmium-metallothioneins. Toxicol Lett 2003;136:193-8. [Crossref] [PubMed]
- Smith DJ, Jaggi M, Zhang W, et al. Metallothioneins and resistance to cisplatin and radiation in prostate cancer. Urology 2006;67:1341-47. [Crossref] [PubMed]
- Yap X, Tan HY, Huang J, et al. Over-expression of metallothionein predicts chemoresistance in breast cancer. J Pathol 2009;217:563-70. [Crossref] [PubMed]
- Zbinden S, Wang J, Adenika R, et al. Metallothionein enhances angiogenesis and arteriogenesis by modulating smooth muscle cell and macrophage function. Arterioscler Thromb Vasc Biol 2010;30:477-82. [Crossref] [PubMed]
- Kojima I, Tanaka T, Inagi R, et al. Metallothionein is upregulated by hypoxia and stabilizes hypoxia-inducible factor in the kidney. Kidney Int 2009;75:268-77. [Crossref] [PubMed]
- Nielsen AE, Bohr A, Penkowa M. The balance between life and death of cells: roles of metallothioneins. Biomarker Insights 2007;1:99-111. [PubMed]
- Dziegiel P, Jeleń M, Muszczyńska B, et al. Role of metallothionein expression in non-small cell lung carcinomas. Rocz Akad Med Bialymst 2004;49:43-5. [PubMed]
- Schmitz KJ, Lang H, Kaiser G, et al. Metallothionein overexpression and its prognostic relevance in intrahepatic cholangiocarcinoma and extrahepatic hilar cholangiocarcinoma (Klatskin tumors). Hum Pathol 2009;40:1706-14. [Crossref] [PubMed]
- Tüzel E, Kirkali Z, Yörükoglu K, et al. Metallothionein expression in renal cell carcinoma: subcellular localization and prognostic significance. J Urol 2001;165:1710-3. [Crossref] [PubMed]
- Surowiak P, Materna V, Maciejczyk A, et al. Nuclear metallothionein expression correlates with cisplatin resistance of ovarian cancer cells and poor clinical outcome. Virchows Arch 2007;450:279-85. [Crossref] [PubMed]
- Weinlich G, Zelger B. Metallothionein overexpression, a highly significant prognostic factor in thin melanoma. Histopathology 2007;51:280-3. [Crossref] [PubMed]
- Królicka A, Kobierzycki C, Puła B, et al. Comparison of metallothionein (MT) and Ki-67 antigen expression in benign and malignant thyroid tumours. Anticancer Res 2010;30:4945-9. [PubMed]
- Kanda M, Nomoto S, Okamura Y, et al. Detection of metallothionein 1G as a methylated tumor suppressor gene in human hepatocellular carcinoma using a novel method of double combination array analysis. Int J Oncol 2009;35:477-83. [PubMed]
- Yan DW, Fan JW, Yu ZH, et al. Downregulation of metallothionein 1F, a putative oncosuppressor, by loss of heterozygosity in colon cancer tissue. Biochimica Et Biophysica Acta 2012;1822:918-26. [Crossref] [PubMed]


