
Upregulated necroptosis-pathway-associated genes are unfavorable prognostic markers in low-grade glioma and glioblastoma multiforme
Introduction
Glioma accounts for ~70% of primary brain malignancies in adults. Historically, the most important prognostic factor for glioma is its histological grade according to the WHO classifications (grade I–IV) (1). While the 2007 WHO classification of central nervous system (CNS) tumors was mainly based on subjective morphology, the current classification updated in 2016 include molecular markers such as IDH mutation, +1p/19q co-deletion and so on, suggesting the significance of biomarkers in prognosis (2).
Necrosis of tumors plays an important role in tumorigenesis as well as response to therapy thus influences prognoses (3). This form of death, was earlier considered as an unregulated process, while a regulated form of necrosis, referred to as “necroptosis” has been recently identified. The main regulators of necroptosis pathway are receptor interacting protein kinasess1 and 3 (RIPK1 and RIPK3) and multiple lineage like kinase (MLKL) (4,5). Upon necroptotic stimuli, RIPK1 recruits RIPK3, which combines to form the necrosome and induce RIPK3 phosphorylation. Subsequently, activated RIPK3 can phosphorylate MLKL, leading to the occurrence of necroptosis. siRNA screen has identified 432 genes that are involved in the regulation of necroptosis (6). Interestingly, most of these differentially expressed genes are enriched in nervous system and immune system, suggesting a close relationship between necroptosis and nervous system. Currently, the studies investigating relationship between necroptosis and diseases of the nervous system are mainly limited to non-neoplastic conditions such as cerebral ischemia (7), traumatic brain injury (8), Huntington’s disease (9), Alzheimer’s disease (10), and Parkinson’s disease (11). For instance, activation of necroptosis by overexpression of MLKL effectively induces Alzheimer’s disease, while blockage of necroptosis alleviates the disease (10). Moreover, inhibiting necroptosis of oligodendrocyte precursor cells by a necroptosis inhibitor-Necrostatin-1 has been found to reduce white matter damage after cerebral ischemia (7). Reduced RIPK3 has been shown to ameliorate oxidative stress and inflammation protecting astrocytes during traumatic brain injury (8). It is well known that oligodendrocyte precursor cells are the most abundant cell population in human brain which were suspected to be the genesis of gliomas due to cell death disorder (12). Although studies related to the impact of necroptosis on disease process in neuro-oncology are limited, available information suggests a possible link between necroptosis-pathway-associated genes and the clinical behavior of CNS tumors. For example, higher expression of RIPK1 was found to be associated with both worse overall survival (OS) and progression-free survival (PFS) rates in glioblastoma multiforme (GBM), which is attributed to the inhibition of P53 induction (13).
The prognostic value of necroptosis-pathway-associated genes (RIPK1, RIPK3, MLKL) in diffuse glioma including low-grade glioma (LGG, WHO II/III) and GBM (WHO IV) is largely unknown. In the present study, we conducted data mining of The Cancer Genome Atlas (TCGA) database by using Gene Expression Profiling Interactive Analysis (GEPIA) to evaluate the prognostic value of necroptosis-pathway-associated genes in LGG and GBM.
Methods
Different expression of necroptosis-pathway-associated genes between tumor and normal tissue
We used an online tool—GEPIA (http://gepia.cancer-pku.cn/, updated by November 13, 2018) to analyze different expression of RIPK1, RIPK3 and MLKL between tumor and normal tissues in LGG and GBM. The cutoff of |Log2FC| was 1. The cut-off P value was 0.01. TCGA normal and GTEx data were both chosen to match the tumor as normal data.
Correlation between RIPK1, RIPK3 and MLKL genes in LGG and GBM
Correlation analysis was performed to examine the relationship between the three necroptosis-associated genes. Pearson was used to calculate the correlation coefficient.
Survival analysis
The median expression of RIPK1, RIPK3 and MLKL was used to divide patients into high versus low expression groups. Hazards ratio (HR) was calculated based on Cox PH Model on OS and disease-free survival (DFS) with 95% confidence interval. The survival curves were drawn using the GEPIA database.
Results
Differential expression of necroptosis-pathway-associated genes between tumor and normal tissue
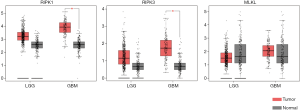
In LGG, the expression levels of RIPK1 and RIPK3 were a marginally upregulated in the tumor tissue compared with the normal tissue, although it was not statistically significant, while the expression of both these genes were significantly higher in GBM as compared to the normal tissue (Figure 1). However, no significant difference was observed in the expression of MLKL in both LGG and GBM (Figure 1).
Correlation between RIPK1, RIPK3 and MLKL genes in LGG and GBM
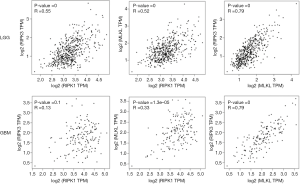
We also analyzed the correlation among these three different genes and found a moderate correlation between MLKL and RIPK3 in both LGG (R =0.79) and GBM (R =0.79) (Figure 2). In contrast, the correlation between RIPK3 and RIPK1 as well as RIPK1and MLKL was rather weak.
RIPK1, RIPK3, and MLKL genes were associated with OS and DFS in LGG
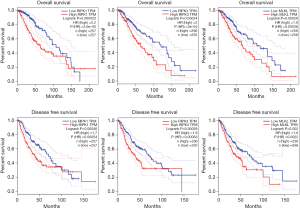
In LGG patients, the expressions of RIPK1, RIPK3, and MLKL correlated negatively with OS, thus higher expression levels of these three regulators was related to poor survival. The corresponding HR values were found to be 2.2, 2 and 1.9 respectively (P=0.000023, 0.00024 and 0.00029). Similar observations were made in case of DFS, where higher expression levels of RIPK1, RIPK3, and MLKL was correlated with poor DFS, and the HR values were 1.7, 1.8 and 1.6 respectively (P=0.00046, 0.00028 and 0.002). Further, differences in OS and DFS between high and low expression groups of all three genes were found to be statistically significant (Figure 3).
MLKL gene was associated with OS and DFS in GBM
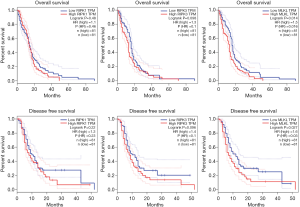
In GBM, the expression level of MLKL correlated negatively with OS and DFS, while such correlation could not be observed in case of RIPK1 and RIPK3 (Figure 2). Therefore, higher levels of MLKL appear to be related to worse OS and DFS with HR values of 1.5 and 1.6 respectively (P=0.014, 0.027). The corresponding HR values for OS and DFS were also rather low for RIPK1 (1.1 and 1.3; respectively; P=0.48, 0.22), and RIPK3 (1.3 and 1.4; P=0.098, 0.096, respectively) (Figure 4).
Discussion
We analyzed the relationship between necroptosis-pathway-associated genes and prognosis in LGG and GBM by using Gepia, a platform which has been established to facilitate bioinformatics analysis of data from TCGA (14). We found that RIPK1, RIPK3, and MLKL can serve as a prognostic marker for both LGG and GBM, with higher expression of these necroptosis-pathway-associated genes as a negative prognosticator. To our knowledge, this is the first study investigating the potential of necroptosis-pathway-associated genes as prognostic markers in both LGG and GBM.
Earlier studies have suggested that higher levels of expression of necroptosis-associated genes could be related to favorable prognoses in several malignancies. For instance, restoration of RIP3 expression using hypomethylating agents has been reported to increase in chemosensitivity and enhanced metastatic relapse-free survival in breast cancer patients with higher RIP3 expression (15). A positive correlation between higher MLKL expressions has been reported in a subset of patients receiving adjuvant treatment following resection in pancreatic adenocarcinoma (16). In ovarian cancer, higher MLKL expression has been found to be associated with better DFS and OS as compared to patients with lower MLKL expression (17). However, studies on gastric, esophageal cancer and colorectal cancers suggest that necroptosis-pathway-associated genes are negatively correlated with patients’ survival (18,19). Thus, it appears that the relationship between necroptosis-pathway-associated genes and prognosis may depend on the type of the malignancy as well as clinical scenario.
Results of in vitro (established cell lines) and animal studies have provided evidences for the dual role for necroptotic signaling in cancer. On the one hand, activated necroptosis-pathway can lead to immune-stimulation, inducing the release of damage-associated molecular pattern (DAMPs) such as high-mobility group box-1 (HMGB1), calreticulin and ATP from necroptotic tumor cells, which can stimulate antigen presenting cells of the innate immune system to migrate to lymphoid tissues and function as immunostimulatory factors for T cells (20). On the other hand, necroptosis-pathway also leads to immunosuppression in the microenvironment favoring oncogenesis (21). Collectively, these findings suggest a complex role of necroptosis machinery in cancer, which appears to be context-dependent, thus requiring additional information in managing the clinical scenario.
Our analysis demonstrated that up-regulation of necroptosis-associated genes was related to worse OS and DFS in LGG patients, while only a higher expression of MLKL was related to worse OS and DFS in GBM patients. Although an earlier study showed that in GBM, a higher level of RIPK1 protein negatively correlated with prognosis (13), such a prognostic value of RIPK1 gene expression was not observed in the present study (Figure 4), suggesting possible differences between the expression and protein levels of this gene. Despite these differences, both studies showed that higher necroptosis-pathway signaling is a negative prognostic biomarker in GBM. The underlying mechanisms for such observation are lacking and needs to be unraveled. It is noteworthy that brain was generally recognized as an immune privileged organ because of the blood-brain barrier, which may limit the immunostimulation by the necroptosis pathway. Moreover, HMGB1, generally considered as a potent inducer of immunogenic cell death, has also been found to enhance inflammation, proliferation and migration of GBM cells in vitro (22,23). Taken together, the present study demonstrates the prognostic value of necroptosis-associated genes in LGG and GBM, while the underlying mechanism needs to be explored in the future.
There are certain limitations of the present study that requires to be mentioned. Firstly, GEPIA at present provide only RNA sequencing data (gene expression), but not the protein level or necroptosis-specific phosphorylation level of RIPK1, RIPK3, MLKL, which are the hallmarks of the activated necroptosis pathway. Secondly, the reason why upregulated necroptosis-pathway-associated genes are associated with unfavorable prognosis in LGG and GBM remain to be explored. In future studies, we plan to validate our observations in cell or animal experiments.
Conclusions
The present study suggests that up-regulated necroptosis-pathway-associated genes are associated with unfavorable prognosis in patients with LGG and GBM. These findings provide new insight into the potential role of necroptosis-pathway-associated genes in future clinical application.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.05.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ricard D, Idbaih A, Ducray F, et al. Primary brain tumours in adults. Lancet 2012;379:1984-96. [Crossref] [PubMed]
- Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007;114:97-109. [Crossref] [PubMed]
- Richards CH, Mohammed Z, Qayyum T, et al. The prognostic value of histological tumor necrosis in solid organ malignant disease: a systematic review. Future Oncol 2011;7:1223-35. [Crossref] [PubMed]
- Sun L, Wang H, Wang Z, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012;148:213-27. [Crossref] [PubMed]
- Chan FK, Luz NF, Moriwaki K. Programmed necrosis in the cross talk of cell death and inflammation. Annu Rev Immunol 2015;33:79-106. [Crossref] [PubMed]
- Hitomi J, Christofferson DE, Ng A, et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 2008;135:1311-23. [Crossref] [PubMed]
- Chen Y, Zhang L, Yu H, et al. Necrostatin-1 Improves Long-term Functional Recovery Through Protecting Oligodendrocyte Precursor Cells After Transient Focal Cerebral Ischemia in Mice. Neuroscience 2018;371:229-41. [Crossref] [PubMed]
- Liu ZM, Chen QX, Chen ZB, et al. RIP3 deficiency protects against traumatic brain injury (TBI) through suppressing oxidative stress, inflammation and apoptosis: Dependent on AMPK pathway. Biochem Biophys Res Commun 2018;499:112-9. [Crossref] [PubMed]
- Vandenabeele P, Galluzzi L, Vanden Berghe T, et al. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 2010;11:700-14. [Crossref] [PubMed]
- Caccamo A, Branca C, Piras IS, et al. Necroptosis activation in Alzheimer's disease. Nat Neurosci 2017;20:1236-46. [Crossref] [PubMed]
- Iannielli A, Bido S, Folladori L, et al. Pharmacological Inhibition of Necroptosis Protects from Dopaminergic Neuronal Cell Death in Parkinson's Disease Models. Cell Rep 2018;22:2066-79. [Crossref] [PubMed]
- Dufour A, Gontran E, Deroulers C, et al. Modeling the dynamics of oligodendrocyte precursor cells and the genesis of gliomas. PLoS Comput Biol 2018;14:e1005977. [Crossref] [PubMed]
- Park S, Hatanpaa KJ, Xie Y, et al. The receptor interacting protein 1 inhibits p53 induction through NF-κB activation and confers a worse prognosis in glioblastoma. Cancer Res 2009;69:2809-16. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102. [Crossref] [PubMed]
- Koo GB, Morgan MJ, Lee DG, et al. Methylation-dependent loss of RIP3 expression in cancer represses programmed necrosis in response to chemotherapeutics. Cell Res 2015;25:707-25. [Crossref] [PubMed]
- Colbert LE, Fisher SB, Hardy CW, et al. Pronecrotic mixed lineage kinase domain‐like protein expression is a prognostic biomarker in patients with early‐stage resected pancreatic adenocarcinoma. Cancer 2013;119:3148-55. [Crossref] [PubMed]
- He L, Peng K, Liu Y, et al. Low expression of mixed lineage kinase domain-like protein is associated with poor prognosis in ovarian cancer patients. Onco Targets Ther 2013;6:1539-43. [PubMed]
- Zhu G, Ye J, Huang Y, et al. Receptor-interacting protein-1 promotes the growth and invasion in gastric cancer. Int J Oncol 2016;48:2387-98. [Crossref] [PubMed]
- Liu X, Zhou M, Mei L, et al. Key roles of necroptotic factors in promoting tumor growth. Oncotarget 2016;7:22219-33. [PubMed]
- Galluzzi L, Buqué A, Kepp O, et al. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol 2017;17:97-111. [Crossref] [PubMed]
- Seifert L, Werba G, Tiwari S, et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature 2016;532:245-9. [Crossref] [PubMed]
- Bassi R, Giussani P, Anelli V, et al. HMGB1 as an autocrine stimulus in human T98G glioblastoma cells: role in cell growth and migration. J Neurooncol 2008;87:23-33. [Crossref] [PubMed]
- Zhang J, Liu C, Hou R. Knockdown of HMGB1 improves apoptosis and suppresses proliferation and invasion of glioma cells. Chin J Cancer Res 2014;26:658-68. [PubMed]


