
Identification of high expression profiles of miR-31-5p and its vital role in lung squamous cell carcinoma: a survey based on qRT-PCR and bioinformatics analysis
Introduction
Lung and bronchus cancer is the principal cause of death and the second highest incidence rate among all malignancies worldwide. Studies show that lung cancer causes exceed one-quarter deaths from cancers in the US (1,2). Lung cancer include two major histological categories: small cell lung cancer (SCLC 15%) and non-small cell lung cancer (NSCLC 85%). NSCLC can be divided into three types, include lung large cell carcinoma (LULC), lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) (3-5). With the rapid growth of incidence of lung cancer, more and more studies have been made about its occurrence, development and metastatic mechanism in recent years, however, few studies focused on the molecular mechanism of LUSC.
By degrading mRNAs or inhibiting translation, MiRNAs could induce gene silencing (6). Researches confirm that MiRNAs play an important role in proliferation, growth, invasion and apoptosis of cancers (7-10). For example, studies identified that miR-132, miR-198 and miR-1269 may indicate the occurrence, development and prognosis of hepatocellular carcinoma through their potential molecular mechanism and their targeted genes Zhang et al. suggested that downregulation of miR-132 may result from the HBx expression via DNA methylation, study from Huang et al. reported that through targeting fibroblast growth factor receptor 1, miR-198 could decrease proliferation and depress apoptosis, Gan et al. reported that DACT1 may be target gene of miR-1269,and downed-regulated expression of DACT1 may result in infiltration and metastasis of tumor by inhibiting NF-κB signaling and WNT/beta-catenin signaling pathways (11-13). Some researches have also reported the important value of miRNAs in the diagnosis, therapy, and prognosis of LUSC (14-16) . Nevertheless, the role of miR-31-5p in lung squamous cell carcinoma (LUSC) was unknown (17) Therefore, additional studies are needed to clarify the molecular mechanism of miR-31-5p in LUSC.
Previous studies reported that the expression of miR-31-5p was up-regulated in pancreatic cancer, colorectal cancer and hepatocellular carcinoma (18-20), but it was down-regulated in cemento-ossifying fibroma and upper tract urothelial carcinoma (21,22). Nevertheless, only a few reports about the relationship between miR-31-5p and NSCLC have been published up to now. Chun et al. reported a high expression of miR-31-5p in NSCLC tissue (23), but this study was limited by the quantity of the tissue samples and different methods of data collection and analysis. In this study, we intend to detect the expression of miR-31-5p in LUSC and confirm the miR-31-5p target genes by using several online databases.
More studies were needed to understand the relationship between miR-31-5p and LUSC. In current study, we intend to detect the expression of miR-31-5p in LUSC and predict the target genes of miR-31-5p by several online databases. We will also analyze their molecular mechanisms by bioinformatics software and analyze which function and signaling pathway they have impact, expect to provide theoretical foundation for finding new therapeutic targets of LUSC.
Methods
Tissue samples
Our research gained the ethics approval from The Medical Ethics Committee of First Affiliated Hospital of Guangxi Medical University [approval number:2019 (K-E-Y-019)]. A total of 88 LUSC patients (Ages range from 23 to 82, with an average age of 56.5, include 50 males and 38 females) who had been treated in the First Affiliated Hospital of Guangxi Medical University were chosen for present study. All tissues were formalin fixed paraffin embedded (FFPE). All the clinical features were exhibited in Table 1.
Table 1
| Clinicopathological parameters | n | Relevant expression of miR-31-5p (2-ÄCq) | ||
|---|---|---|---|---|
| Mean ± SD | t | P value | ||
| Tissue | 7.083 | 0.000 | ||
| Para-carcinoma tissue | 88 | 4.3008±1.73832 | ||
| LUSC | 88 | 6.1472±1.71976 | ||
| Age (years) | 0.119 | 0.905 | ||
| <60 | 51 | 6.1659±1.67066 | ||
| ≥60 | 37 | 6.1214±1.80824 | ||
| Gender | 0.086 | 0.932 | ||
| Male | 50 | 6.1610±1.88261 | ||
| Female | 38 | 6.1289±1.50332 | ||
| Smoke | −0.603 | 0.548 | ||
| No | 33 | 6.2903±2.03028 | ||
| Yes | 55 | 6.0613±1.51732 | ||
| Tumor size (cm) | −0.915 | 0.590 | ||
| ≤3 | 49 | 6.4557±1.82857 | ||
| >3 | 39 | 5.7595±1.50688 | ||
| Lymph node metastasis | −1.140 | 0.257 | ||
| No | 46 | 6.3465±1.76204 | ||
| Yes | 42 | 5.9288±1.66573 | ||
| Vascular invasion | 0.651 | 0.651 | ||
| No | 66 | 6.0780±1.75576 | ||
| Yes | 22 | 6.3545±1.62820 | ||
| TNM | −0.923 | 0.359 | ||
| I–II | 33 | 5.9265±1.60711 | ||
| III–IV | 55 | 6.2784±1.78540 | ||
LUSC, lung squamous cell carcinoma.
Quantitative real-time reverse transcription PCR (qRT-PCR)
qRT-PCR was performed to detect the expression of miR-31-5p in 88 FFPE tissues as a previous study reported (24). Additionally, using NormFinder and geNorm, we found that the miR-191 and miR-103 were the most stable housekeeping miRNAs, so they were selected as the endogenous controls. The sequences of the miRNAs are as follows: miR-191(Applied Biosystems Cat. No. 4427975-000490): CAACGGAAUCCCAAAAGCAGCU; miR-103 (Applied Biosystems Cat. No. 4427975-000439): AGCAGCAUUGUACAGGGCUAUGA; and miR-31-5p (AGGCAAGAUGCUGGCAUAGCU). Applied Biosystems PCR7900 (Foster City, CA, USA) was adopted to perform the quantitative real-time polymerase chain reaction (qRT-PCR). The formula2-ΔCq was utilized to determine the expression level of miR-31-5p (25).
Information extraction of miR-31-5p in LUSC from GEO and TCGA datasets
The Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) (26) was used for extracting the expression information of miR-31-5p in LUSC. Appropriate data were screened and selected on the basis of the criteria listed below. The inclusion criterias of this study were as follows: (I) Studies had examined the expression level of miR-31-5p in blood or tissue samples; (II) Human species that were diagnosed with lung squamous cell carcinoma; (III) Both tumor specimens and normal specimens original expression data of miR-31-5p was provided. Exclusion criterias of the current study were as follows: (I) Animal or cell line research data; (II) Tumor or normal groups with small sample sizes (n<10). We also extracted the information from TCGA data portal (http://tcga-data.nci.nch.gov/tcga). Then, these data were summarized, calculated and analyzed using SPSS 22.0 (SPSS, IBM, USA). The primary expression data were log-transformed when they haven’t been normalized.
A meta-analysis based on data in our current study, together with data from the GEO, and TCGA, was conducted in our study. In addition, receiver operating characteristic (ROC) curve was made to confirm the different expression of miR-31-5p based on each dataset. Meanwhile, summary receiver operating characteristic (sROC) curve was also applied for further confirmation of the different expression levels of miR-31-5p between LUSC tissues and the normal tissues.
Prediction of miR-31-5p potential target genes
The target genes of miR-31-5p were determined using the following 12 online bioinformatics software: miRWalk, MicroT4, RNA22, RNAhybrid, miRanda, miRBridge, miRNAMap, PICTAR2, PITA, miRDB, miRMap and Targetscan. The genes present in more than 6 prediction software packages were selected as the potential target genes of miR-31-5p. We also searched for the target genes of miR-31-5p in PubMed and EMBASE. Meanwhile, genes that were confirmed by experiments were accepted. For higher specificity, we chose the intersection elements of potential genes and the down-expression genes as the target genes of miR-31-5p.
Bioinformatics analysis of the potential target genes of miR-31-5p
The Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) was used for GO, KEGG and analysis of those potential genes. GO analysis investigates the pathways from the cell component (CC), molecular function (MF), and biological processes (BP), and KEGG was applied to determine the remarkable pathways related to those genes.
PPI network construction
The PPI network, which was based on the STRING 10.5 online software (https://string-db.org/), was utilized to point out the association of those target genes. At the same time, the hub genes (with the largest connections in the PPI network) that may play an important part in the strategic pathway of LUSC have been identified.
Statistical analyses
Statistical Package for the Social Sciences (SPSS, IBM, USA) 22.0, GraphPad Prism 5 (Inc., La Jolla, CA, USA), StataSE 12.0, and R version 3.3.0 were utilized for all of the statistical analyses. The scatter diagram was applied to show the different expressions of miR-31-5p between patients with LUSC and the controls. Meanwhile, receiver operating characteristic (ROC) and the area under the curve (AUC) were applied to confirm the differential expression of miR-31-5p in patients with LUSC and corresponding controls. Possible correlations between miR-31-5p expression and clinical features were examined using the Spearman correlation. StataSE 12.0 was used for the meta-analysis of the data and SMD <0 and 95% CI did not cross zero represented miR-31-5p had a lower expression in LUSC tissues than in normal specimens. In contrast, in this case, when the overall SMD >0 and 95% CI did not cross zero, tumor specimens were supposed to have a higher expression of miR-31-5p than normal specimens. Moreover, funnel plot and Egger’s test were utilized for detecting the publication bias. In all the analyses, the P value <0.05 was considered to be statistically significant.
Results
High expression of miR-31-5p in LUSC
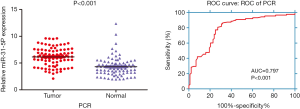
The expression data of miR-31-5p in 88 LUSC tissues and the controls was detected by qRT-PCR, the result showed that the expression of miR-31-5p was significantly higher in LUSC than the normal lung tissues (P<0.001), as shown by the AUC of 0.797 (Figure 1).
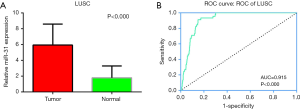
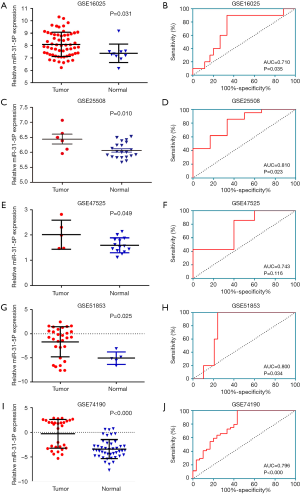
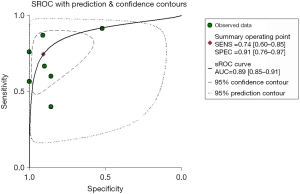
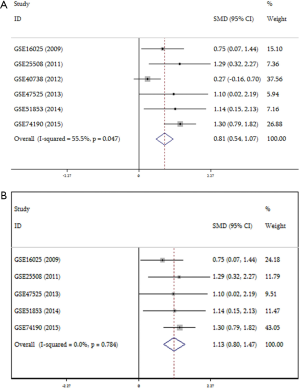
Meanwhile, data from TCGA showed the expression of miR-31-5p in LUSC was significantly higher than the controls (P<0.001) (Figure 2A) and the AUC was 0.915 (CI:1.197–1.624, P<0.000) (Figure 2B Respect to the GEO database, five databases (GSE16025, GSE25508, GSE47525, GSE51853, GSE74190) supported the viewpoint that the up-regulated expression of miR-31-5p in LUSC patients compared to the corresponding controls (P<0.05) (Figure 3). Furthermore, the ROC curves, which were based on the data taken from 6 GEO datasets, were utilized for confirming the high expression levels of miR-31-5p in LUSC (shown in Figure 3B,D,F,G,H), reveal that five of the six datasets from GEO support the high expression of miR-31-5p in LUSC, including GSE51858 (AUC =0.800, P=0.034), GSE74190 (AUC =0.796, P<0.000), GSE47525 (AUC =0.743, P=0.116), GSE16025 (AUC =0.710, P=0.035). The ROCs and AUCs also proved the higher expression of miR-31-5p in LUSC [GSE51858 (AUC =0.800, P=0.034), GSE74190 (AUC =0.796, P<0.000), GSE16025 (AUC =0.710, P=0.035), GSE25508 (AUC =0.810, P=0.023), and GSE47525 (P=0.049, AUC =0.743)] and GSE25508 (AUC =0.810, P=0.023), but the other one was without statistical significance (P>0.05, figure not shown). Meanwhile, the SROC which was based on five GEO database, TCGA and our study, also supported the up-regulated expression of miR-31-5p in LUSC (AUC=0.89) (Figure 4).
Meta-analysis of miR-31-5p expression in LUSC
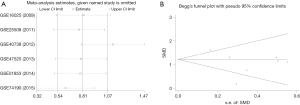
A total of 1,012 samples, including 719 tumor and 283 normal samples, of which 313 cases came from 6 GEO datasets, 523 cases were obtained from the TCGA dataset and 176 samples from our research, were sent to StataSE for meta-analysis. According to the data from the six GEO datasets, the forest plot showed heterogeneity is exist (I2 =55.5%, Figure 5). Then, sensitivity analysis of the forest study implied that data from GSE40738 may be heterogeneous in our study (P=0.285, Figure 6A), and another forest plot without GSE40738 dataset was conducted, which showed no heterogeneity in those five GEO datasets (I2=0.00%, Figure 5B). In addition, it showed higher expression of miR-31-5p in LUSC tissues compared to normal tissues (SMD =1.132, CI: 0.797–1.467, Z=6.62, P=0.000). Furthermore, the funnel plot showed that there was no publication bias in the studies (P=0.285, Figure 6B).
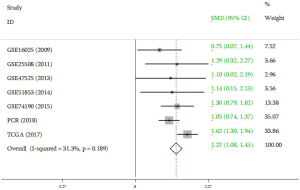
Meanwhile, the meta-analysis result of all the data in the current study (PCR, TCGA, GEO included) showed that the expression of miR-31-5p significantly increased in patients with LUSC than the controls (SMD =0, 95% CI: 1.08–1.45, Z=13.30, P=0.000, Figure 7).
Potential target genes of miR-31-5p
In total, 15,947 genes were screened base on 12 online predict softwares, and the genes present on more than 6 programs were selected to overlap with genes that were down-regulated in LUSC. Finally, 316 overlapped genes and 6 genes that were validated by the document were chosen as the target genes of miR-31-5p.
GO and KEGG pathway analyses
A total of 322 potential target genes of miR-31-5p were subjected to GO and KEGG analysis. As shown by GO analysis, these genes showed a great effect on signal transduction (GO: 0007165, P<0.001), positive regulation of endothelial cell proliferation (GO: 0001938, P<0.001) and peptidyl-tyrosine phosphorylation (GO: 0018108, P<0.001) for BP. On the level of MF, they were significantly associated with transmembrane receptor protein tyrosine kinase activity (GO: 0004714, P<0.001), Ras guanyl-nucleotide exchange factor activity (P<0.001, GO: 0005088) and growth factor binding (GO: 0005088, P<0.001) (Table 2). With respect to CC, the terms that most relate to the target genes were plasma membrane (GO: 0005886, P<0.001), integral component of the plasma membrane (GO: 0005887, P<0.001) and membrane raft (GO: 0045121, P<0.001). The KEGG analysis revealed the top three signaling pathways were Rap1 signaling pathway (hsa04015, P<0.05), Oxytocin signaling pathway (hsa04921, P<0.05) and Proteoglycans in cancer (hsa05205, P<0.05) (Table 3).
Table 2
| Category | Term | Count | P value |
|---|---|---|---|
| KEGG_PATHWAY | Rap1 signaling pathway | 14 | 1.10×10-4 |
| KEGG_PATHWAY | Oxytocin signaling pathway | 11 | 5.80×10-4 |
| KEGG_PATHWAY | Proteoglycans in cancer | 12 | 1.00×10-3 |
| KEGG_PATHWAY | Melanogenesis | 8 | 1.00×10-3 |
| KEGG_PATHWAY | Tight junction | 8 | 2.20×10-3 |
Table 3
| Category | Terms | Counts | P value |
|---|---|---|---|
| GOTERM_BP_DIRECT | Signal transduction | 45 | 4.10×10-7 |
| GOTERM_BP_DIRECT | Positive regulation of endothelial cell proliferation | 9 | 2.10×10-5 |
| GOTERM_BP_DIRECT | Peptidyl-tyrosine phosphorylation | 12 | 6.10×10-5 |
| GOTERM_BP_DIRECT | Positive regulation of GTPase activity | 24 | 1.00×10-4 |
| GOTERM_BP_DIRECT | Positive regulation of cell migration | 12 | 3.10×10-4 |
| GOTERM_BP_DIRECT | Activation of phospholipase C activity | 5 | 9.00×10-4 |
| GOTERM_BP_DIRECT | Transmembrane receptor protein tyrosine kinase signaling pathway | 8 | 1.20×10-3 |
| GOTERM_BP_DIRECT | Intracellular signal transduction | 17 | 1.50×10-3 |
| GOTERM_BP_DIRECT | Adenylate cyclase-activating G-protein coupled receptor signaling pathway | 6 | 1.50×10-3 |
| GOTERM_BP_DIRECT | Establishment of endothelial barrier | 4 | 2.30×10-3 |
| GOTERM_MF_DIRECT | Transmembrane receptor protein tyrosine kinase activity | 7 | 3.40×10-5 |
| GOTERM_MF_DIRECT | Ras guanyl-nucleotide exchange factor activity | 9 | 6.30×10-4 |
| GOTERM_MF_DIRECT | Growth factor binding | 5 | 9.40×10-4 |
| GOTERM_MF_DIRECT | Rho guanyl-nucleotide exchange factor activity | 7 | 1.70×10-3 |
| GOTERM_MF_DIRECT | Steroid hormone receptor activity | 6 | 2.20×10-3 |
| GOTERM_CC_DIRECT | Plasma membrane | 113 | 1.10×10-8 |
| GOTERM_CC_DIRECT | Integral component of plasma membrane | 54 | 2.00×10-8 |
| GOTERM_CC_DIRECT | Membrane raft | 17 | 3.90×10-7 |
| GOTERM_CC_DIRECT | Cell surface | 24 | 8.20×10-4 |
| GOTERM_CC_DIRECT | Focal adhesion | 17 | 9.20×10-4 |
GO, gene ontology.
PPI network analysis for the selection of hub genes.
The potential genes were imported to STRING for association analysis and the result showed 328 nodes and 437 edges among the network that was made up of 322 potential target genes of miR-31-5p (Figure 8A). Among these, the 6 hub genes of miR-31-5p, including adenylate cyclase 9 (ADCY9), adenylate cyclase 6 (ADCY6), neuromedin U receptor 1 (NMUR1), C-X-C motif chemokine ligand 12 (CXCL12), signal transducer and activator of transcription 5A (STAT5A) and epidermal growth factor receptor (EGFR) (Figure 8B).
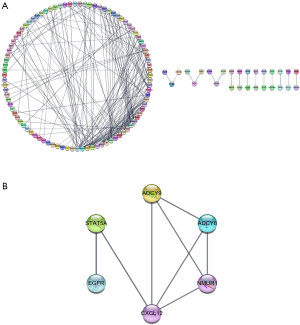
Three of these hub genes, including ADCY6, ADCY9 and EGFR, appeared on the Rap1 signaling pathway, Oxytocin signaling pathway and Melanogenesis simultaneously.
Discussion
The poor diagnosis of the early stage disease and the useless treatment methods leads to poor prognosis of LUSC. Therefore, it is important to find an effective way to diagnose and treat lung cancer. In the current study, we found that the expression profile of miR-31-5p was up-expressed in LUSC tissue compared to the normal tissue. We also confirmed the high miR-31-5p expression in LUSC through the ROC and sROC curve. Meanwhile, 322 potential targeted genes of miR-31-5p were predicted, and the molecular pathways like signal transduction, and Rap1 signaling pathway and transmembrane receptor protein tyrosine kinase activity, that may be involved in LUSC were discovered. Nevertheless, 6 hub genes of miR-31-5p were chosen for exploration of the molecular mechanism that involved in LUSC, among which, three genes were found to coexist in three pathways. These findings indicate that miR-31-5p play a major role in the initiation and progression of LUSC.
Studies improved that miR-31-5p closely related to the occurrence and development of tumors. Zhong et al. found that knockdown of miR-31-5p resulted in increasing expression of hMLH1 protein and reducing cell cycle arrest in NSCLC cells. They also reported that overexpression of miR-31-5p lead to decrease hMLH1 protein expression and induced cell cycle arrest at S phase (27). Mlcochova et al. (28) reported down-regulation of miR-31-5p in metastatic colorectal cancer (mCRC), and found it influenced the predicting the time to progression (TPP) in patients who received cetuximab treatment, and after transfecting several cell lines (HCT-116, DLD-1 and HT-29) in mCRC, 84 upregulated genes and 148 downregulated genes came to light (P<0.01) and Slattery et al. also reported the correlation between colorectal tumor molecular phenotype and miR-31-5p, and they found that miR-31-5p is significantly related to the KRAS and BRAF mutations, which are associated with survival of patients with the colorectal tumor molecular phenotype (29). Capri et al. reported that miR-31-5p had four seed sequences within the 30 UTR of GLT1 (30). ZHANG G validated another target gene (SP1) of miR-31-5p and suggested that up-regulated expression of miR-31-5p came into play as a suppressor in hepatocellular carcinoma (18). Accumulating evidence shows that miR-31-5p is expressed differently in different cancers, which affects the prognosis, diagnosis and treatment of cancers, until some molecular mechanisms have been confirmed.
In the current study, we focus on finding the potential signal pathways that are related to miR-31-5p in LUSC. Our results, 25 potential pathways that supposedly correlate with miR-31-5p are displayed in the Tables 1-3. In addition, the protein interaction network based on those potential target genes also has been conducted, and we then selected the top six genes for further study. This study intended to find the function of the six potential genes of miR-31-5p, to better understand the molecular mechanism in LUSC. Here, we would like to discuss the possible mechanisms of the genes that were verified by previous studies.
C-X-C motif chemokine ligand 12 (CXCL12) was proven by a number of studies to be connected with tumor growth, invasion, angiogenesis, and metastasis in different types of cancer, such as epithelial ovarian carcinoma, breast cancer, clear cell renal cell carcinoma, prostate cancer and pancreatic cancer. As a result, researchers identified the connection between CXCL12 and diagnosis, prognosis and treatment of cancers in patients (31-34). Wang et al. reported that up-regulation of CXCL12 expression enhances lung cancer progression through the CXCR4/JAK2 pathway (35). Another study showed CXCL12 expression was connected with a reduced overall survival in lung cancer patients (36). Wang et al. suggested that through activating cancer stem cell, CXCL12 could influenced initiation and progression of lung cancer (37). However, the role that CXCL12 played in LUSC remains unclear.
STAT5A, an encoded member of the STAT family, was identified to be a downstream target of miR-1469. The miR-1469 can bind the 3'-untranslated region of the Stat5a directly, and the expression of mRNA and protein of Stat5a was reduced. Finally, it was proved that by targeting STAT5A, the miR-1469 can lead to apoptosis of lung cancer cells (38). Pastuszak-Lewandoska D also found the expression of STAT5A is down-regulated in non-small cell lung cancer (NSCLC), but the mechanism of how it works in LUSC has not been explored (39).
Epidermal growth factor receptor (EGFR), one of the most popular research genes, has been confirmed to have an important function in various types of cancers (40-42). Because EGFR mutation was found in lung squamous cell carcinoma, drugs for lung cancer with EGFR mutation are in clinical trial and show a promising future as a target for lung cancer patients (43)..Liu et al. also found that EGFR-tyrosine kinase inhibitors (EGFR-TKIs) could make a difference for LUSC patients with EGFR mutation, which predicts a better outcome for the patient who is given EGFR-TKIs (44,45).. Meanwhile, Du et al. suggested that EGFR was one of the cross-talk genes that was associated with the 12 pathways that were connected to LUSC (46). On the basis of clinical data analysis, there was significantly correlation between EGFR and the survival of LUSC patients. In conclusion, EGFR plays a significant role in the treatment and survival of patients in LUSC, but the relationship between miR-31-5p and EGFR remains unexplored.
However, there were also quite a few limitations in this study. First, the total 882 samples from GEO, TCGA and the First Affiliated Hospital of Guangxi Medical University underwent different methods of detection, were from different countries and regions, represented different ages of patients and different stages of tumors and a number of other factors, which means additional subgroup analyses within a large sample are needed to search for the source the heterogeneity. Second, miRNAs come into play through different pathways and molecular mechanisms, therefore several bioinformatics methods were used to evaluate the biomolecular function of the targeted genes of miR-31-5p, including GO and KEGG pathway analysis. Beyond that, a PPI network model was used to visually display the connection of those targeted genes. Nevertheless, to confirm the treatment target of LUSC, more verification experiments are indispensable.
In conclusion, higher expression levels of miR-31-5p in LUSC patients than normal were confirmed by meta-analysis based on the data from GEO, TCGA and our experiment. Furthermore, the diagnostic value of miR-31-5p was verified in the current study. In addition, our research proposed that miR-31-5p may play a major role in the carcinogenesis of LUSC by targeting the hub genes we identified. Moreover, the underlying pathways reveal the molecular mechanism of miR-31-5p in LUSC. To conclude, it could be hypothesized that miR-31-5p serves as a molecular biomarker for LUSC in the clinical setting. However, more studies are needed to confirm the actual function of miR-31-5p in diagnosis, prognosis and therapy of LUSC.
Acknowledgments
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The study was funded by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.04.21). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all patients. Our research gained the ethics approval from The Medical Ethics Committee of First Affiliated Hospital of Guangxi Medical University [approval number:2019 (K-E-Y-019)].
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Zhang Y, He RQ, Dang YW, et al. Comprehensive analysis of the long noncoding RNA HOXA11-AS gene interaction regulatory network in NSCLC cells. Cancer Cell Int 2016;16:89. [Crossref] [PubMed]
- Miao Z, Ali A, Hu L, et al. Microtubule actin cross-linking factor 1, a novel potential target in cancer. Cancer Sci 2017;108:1953-8. [Crossref] [PubMed]
- Long F, Su JH, Liang B, et al. Identification of Gene Biomarkers for Distinguishing Small-Cell Lung Cancer from Non-Small-Cell Lung Cancer Using a Network-Based Approach. BioMed Res Int 2015;2015:685303. [Crossref] [PubMed]
- Zheng H, Zhan Y, Liu S, et al. The roles of tumor-derived exosomes in non-small cell lung cancer and their clinical implications. J Exp Clin Cancer Res 2018;37:226. [Crossref] [PubMed]
- Wang JJ, Li ZF, Li XJ, et al. Effects of microRNA-136 on melanoma cell proliferation, apoptosis, and epithelial-mesenchymal transition by targetting PMEL through the Wnt signaling pathway. Biosci Rep 2017;37: [Crossref] [PubMed]
- Strumidło A, Skiba S, Scott RJ, et al. The potential role of miRNAs in therapy of breast and ovarian cancers associated with BRCA1 mutation. Hered Cancer Clin Pract 2017;15:15. [Crossref] [PubMed]
- Zhou Q, Huang SX, Zhang F, et al. MicroRNAs: A novel potential biomarker for diagnosis and therapy in patients with non-small cell lung cancer. Cell proliferation 2017;50: [Crossref] [PubMed]
- Liu CM, Liang D, Jin J, et al. Research progress on the relationship between zinc deficiency, related microRNAs, and esophageal carcinoma. Thorac Cancer 2017;8:549-57. [Crossref] [PubMed]
- Massillo C, Dalton GN, Farre PL, et al. Implications of microRNA dysregulation in the development of prostate cancer. Reproduction 2017;154:R81-R97. [Crossref] [PubMed]
- Zhang X, Tang W, Li R, et al. Downregulation of microRNA-132 indicates progression in hepatocellular carcinoma. Exp Ther Med 2016;12:2095-101. [Crossref] [PubMed]
- Huang WT, Wang HL, Yang H, et al. Lower expressed miR-198 and its potential targets in hepatocellular carcinoma: a clinicopathological and in silico study. Onco Targets Ther 2016;9:5163-80. [Crossref] [PubMed]
- Gan TQ, Tang RX, He RQ, et al. Upregulated MiR-1269 in hepatocellular carcinoma and its clinical significance. Int J Clin Exp Med 2015;8:714-21. [PubMed]
- Muñoz-Largacha JA, Gower AC, Sridhar P, et al. miRNA profiling of primary lung and head and neck squamous cell carcinomas: Addressing a diagnostic dilemma. J Thorac Cardiovasc Surg 2017;154:714-27. [Crossref] [PubMed]
- Wang Q, Liu S, Zhao X, et al. MiR-372-3p promotes cell growth and metastasis by targeting FGF9 in lung squamous cell carcinoma. Cancer Med 2017;6:1323-30. [Crossref] [PubMed]
- Hou Y, Li L, Ju Y, et al. MiR-101-3p Regulates the Viability of Lung Squamous Carcinoma Cells via Targeting EZH2. J Cell Biochem 2017;118:3142-9. [Crossref] [PubMed]
- Hirsh V. New developments in the treatment of advanced squamous cell lung cancer: focus on afatinib. Onco Targets Ther 2017;10:2513-26. [Crossref] [PubMed]
- Qu K, Zhang X, Lin T, et al. Circulating miRNA-21-5p as a diagnostic biomarker for pancreatic cancer: evidence from comprehensive miRNA expression profiling analysis and clinical validation. Sci Rep 2017;7:1692. [Crossref] [PubMed]
- Kiss I, Mlcochova J, Bortlicek Z, et al. Efficacy and Toxicity of Panitumumab After Progression on Cetuximab and Predictive Value of miR-31-5p in Metastatic Wild-type KRAS Colorectal Cancer Patients. Anticancer Res 2016;36:4955-9. [Crossref] [PubMed]
- Zhao G, Han C, Zhang Z, et al. Increased expression of microRNA-31-5p inhibits cell proliferation, migration, and invasion via regulating Sp1 transcription factor in HepG2 hepatocellular carcinoma cell line. Biochem Biophys Res Commun 2017;490:371-7. [Crossref] [PubMed]
- Pereira TD, Brito JAR, Guimaraes ALS, et al. MicroRNA profiling reveals dysregulated microRNAs and their target gene regulatory networks in cemento-ossifying fibroma. J Oral Pathol Med 2018;47:78-85. [Crossref] [PubMed]
- Izquierdo L, Montalbo R, Ingelmo-Torres M, et al. Prognostic microRNAs in upper tract urothelial carcinoma: multicenter and international validation study. Oncotarget 2017;8:51522-9. [Crossref] [PubMed]
- Li C, Yin Y, Liu X, et al. Non-small cell lung cancer associated microRNA expression signature: integrated bioinformatics analysis, validation and clinical significance. Oncotarget 2017;8:24564-78. [PubMed]
- He RQ, Li XJ, Liang L, et al. The suppressive role of miR-542-5p in NSCLC: the evidence from clinical data and in vivo validation using a chick chorioallantoic membrane model. BMC Cancer 2017;17:655. [Crossref] [PubMed]
- Lan D, Zhang X, He R, et al. MiR-133a is downregulated in non-small cell lung cancer: a study of clinical significance. Eur J Med Res 2015;20:50. [Crossref] [PubMed]
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 2013;41:D991-5. [Crossref] [PubMed]
- Zhong Z, Dong Z, Yang L, et al. MicroRNA-31-5p modulates cell cycle by targeting human mutL homolog 1 in human cancer cells. Tumour Biol 2013;34:1959-65. [Crossref] [PubMed]
- Mlcochova J, Faltejskova-Vychytilova P, Ferracin M, et al. MicroRNA expression profiling identifies miR-31-5p/3p as associated with time to progression in wild-type RAS metastatic colorectal cancer treated with cetuximab. Oncotarget 2015;6:38695-704. [Crossref] [PubMed]
- Slattery ML, Herrick JS, Mullany LE, et al. Colorectal tumor molecular phenotype and miRNA: expression profiles and prognosis. Mod Pathol 2016;29:915-27. [Crossref] [PubMed]
- Capri M, Olivieri F, Lanzarini C, et al. Identification of miR-31-5p, miR-141-3p, miR-200c-3p, and GLT1 as human liver aging markers sensitive to donor-recipient age-mismatch in transplants. Aging Cell 2017;16:262-72. [Crossref] [PubMed]
- Saha A, Ahn S, Blando J, et al. Proinflammatory CXCL12-CXCR4/CXCR7 Signaling Axis Drives Myc-Induced Prostate Cancer in Obese Mice. Cancer Res 2017;77:5158-68. [Crossref] [PubMed]
- Mahjoub MA, Bakhshinejad B, Sadeghizadeh M, et al. Combination treatment with dendrosomal nanocurcumin and doxorubicin improves anticancer effects on breast cancer cells through modulating CXCR4/NF-kappaB/Smo regulatory network. Mol Biol Rep 2017;44:341-51. [Crossref] [PubMed]
- Marona P, Gorka J, Mazurek Z, et al. MCPIP1 Downregulation in Clear Cell Renal Cell Carcinoma Promotes Vascularization and Metastatic Progression. Cancer Res 2017;77:4905-20. [Crossref] [PubMed]
- Sleightholm RL, Neilsen BK, Li J, et al. Emerging roles of the CXCL12/CXCR4 axis in pancreatic cancer progression and therapy. Pharmacol Ther 2017;179:158-70. [Crossref] [PubMed]
- Wang M, Lin T, Wang Y, et al. CXCL12 suppresses cisplatin-induced apoptosis through activation of JAK2/STAT3 signaling in human non-small-cell lung cancer cells. Onco Targets Ther 2017;10:3215-24. [Crossref] [PubMed]
- Samarendra H, Jones K, Petrinic T, et al. A meta-analysis of CXCL12 expression for cancer prognosis. Br J Cancer 2017;117:124-35. [Crossref] [PubMed]
- Wang Z, Sun J, Feng Y, et al. Oncogenic roles and drug target of CXCR4/CXCL12 axis in lung cancer and cancer stem cell. Tumour Biol 2016;37:8515-28. [Crossref] [PubMed]
- Xu C, Zhang L, Li H, et al. MiRNA-1469 promotes lung cancer cells apoptosis through targeting STAT5a. Am J Cancer Res 2015;5:1180-9. [PubMed]
- Pastuszak-Lewandoska D, Domanska D, Czarnecka KH, et al. Expression of STAT5, COX-2 and PIAS3 in correlation with NSCLC histhopathological features. PloS One 2014;9:e104265. [Crossref] [PubMed]
- Bredemeier M, Edimiris P, Mach P, et al. Gene Expression Signatures in Circulating Tumor Cells Correlate with Response to Therapy in Metastatic Breast Cancer. Clin Chem 2017;63:1585-93. [Crossref] [PubMed]
- Zhao K, Wang Q, Wang Y, et al. EGFR/c-myc axis regulates TGFbeta/Hippo/Notch pathway via epigenetic silencing miR-524 in gliomas. Cancer Lett 2017;406:12-21. [Crossref] [PubMed]
- Wang Y, Li Y, Xia L, et al. Continued EGFR-TKI with concurrent radiotherapy to improve time to progression (TTP) in patients with locally progressive non-small cell lung cancer (NSCLC) after front-line EGFR-TKI treatment. Clin Transl Oncol 2018;20:366-73. [Crossref] [PubMed]
- Sanchala D, Bhatt LK, Prabhavalkar KS. Therapeutic approaches for the treatment of epidermal growth factor receptor mutated lung cancer. Curr Cancer Drug Targets 2018;18:773-91. [Crossref] [PubMed]
- Liu Y, Zhang Y, Zhang L, et al. Efficacy of epidermal growth factor receptor-tyrosine kinase inhibitors for lung squamous carcinomas harboring EGFR mutation: A multicenter study and pooled analysis of published reports. Oncotarget 2017;8:49680-8. [PubMed]
- Joshi A, Zanwar S, Noronha V, et al. EGFR mutation in squamous cell carcinoma of the lung: does it carry the same connotation as in adenocarcinomas? Onco Targets Ther 2017;10:1859-63. [Crossref] [PubMed]
- Du J, Zhang L. Pathway deviation-based biomarker and multi-effect target identification in asbestos-related squamous cell carcinoma of the lung. Int J Mol Med 2017;39:579-86. [Crossref] [PubMed]


