
Overexpression of UHRF1 and its potential role in the development of invasive ductal breast cancer validated by integrative bioinformatics and immunohistochemistry analyses
Introduction
Breast cancer has long been a threat to women’s health for its high rates of morbidity and mortality. According to the data from the World Health Organization (WHO), breast cancer is the second most deadly cancer among women in the US (1). Although diagnostic approaches and treatments have improved in recent years, the 5-year survival rate is still less than 25% in breast cancer patients with distant metastasis (2). The pathogenesis of breast cancer is complex and involves multiple factors, including cholesterol metabolism (3), hormone levels (4), and diet (5), among others. Understanding the pathogenesis and seeking for biomarkers is urgently needed for the diagnosis, therapy, and prognosis of breast cancer patients.
Ubiquitin-like PHD and RING finger domain-containing protein 1 (UHRF1) [also named nuclear protein 95 (Np95), or inverted CCAAT box-binding protein of 90 kDa (ICBP90) (6)] plays an essential role in cell proliferation and DNA methylation (7,8). Currently, a plethora of research has demonstrated that UHRF1 participates in the development and progression of many cancers (9). It could inactivate tumor suppressor genes by methylation in many cancers, such as non-small lung carcinoma (9), gastric cancer (10), and endometrial carcinoma (11). High expression of UHRF1 is associated with a poor prognosis in many cancers, including hepatocellular carcinoma (10), pancreatic cancer (11), and bladder cancer (12). Previous research has described a differential role of UHRF1 in breast cancer. Some studies found that UHRF1 inhibits the transcription of MDR1 gene, and enhances the chemotherapeutic effects (13), while other studies found that UHRF1 can act as an oncogene, promoting tumorigenesis and metastasis through complicated mechanisms, such as silencing DNA repair genes and inhibiting apoptosis (14). In the breast cancer cell lines, the mRNA (7,15) and protein (16) expression of UHRF1 was upregulated. The UHRF1 DNA levels in the plasma of breast cancer patients were also increased (17). Clinically, the expression of UHRF1 is gradually increased from normal breast tissue to low-grade breast cancer tissue to high-grade breast cancer tissue (6).
Breast cancer is a heterogeneous disease with many pathological types and the different pathological types have different prognoses (18). It is better to detect the different pathological types of breast cancer separately than it is to do so together. In this study we mainly investigated the expression of UHRF1 in invasive ductal carcinomas of breast cancer, which is the major pathological type of breast cancer. This study aimed to study the role of UHRF1 in invasive ductal carcinoma based on bioinformatics analysis, and to investigate its associations with its clinical pathological characteristics and prognostic significance.
Methods
Gene Expression Omnibus (GEO) data analysis
To identify the critical candidate genes in the development and progression of breast cancer, all breast cancer datasets were collected and assembled from the GEO database. The search strategy was formulated was as follows: (malignan* OR cancer OR tumor OR tumor OR neoplas* OR carcinoma) AND (breast OR mammary). Inclusion criteria included the following: (I) the dataset sample organism was from Homo sapiens; (II) both breast cancer and normal breast tissue with more than two samples were included in the datasets; and (III) provided enough mRNA expression data of normal breast tissue or breast cancer were provided for analysis. Meanwhile, exclusion criteria included the following: (I) data based on cell lines; (II) breast cancer or normal breast tissue treated with some specific treatments such as chemotherapeutic drugs, gene knockout etc.; (III) non Homo sapiens test subjects; and (IV) breast cancer patients with other coincident cancers. The quality-controlled and standardized chip data used in the chosen databases were analyzed by Bayesian Analysis performed by R (version: 2.15.3). Probes without complete gene expression data were filtered, and the probes were transformed manually into gene symbols according to GPL570 manually. Genes with an adjusted P value <0.01 and log2-transformed expression fold change >1.5 or <−1.5 were considered differentially expressing genes (DEG) and chosen for further analysis. In this study, GSE10780, which was submitted by Chen et al. (19) to the GEO database and based on the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array) was chosen for analysis. The GSE10780 included 143 histologically normal breast tissues and 42 invasive ductal carcinoma (IDC) tissues, and was analyzed using the Naive Bayes methods after initial quality control, background correction, and standardization. The top 100 DEGs in normal breast tissues and IDC tissues were used to drawn a heatmap by using R (version: 2.15.3). The GSE29044, which was submitted by Colak et al. (20) and was based on GPL570 platform, contained 36 IDC tissue and 67 normal breast tissue, and were analyzed by the same method to validate the results.
The Cancer Genome Atlas (TCGA) data analysis
For cross-validating mRNA overexpression of target genes, Counts were downloaded and extracted from TCGA (http://cancergenome.nih.gov/) during April 2018 by using the data transfer tool (downloaded from https://gdc.cancer.gov/access-data/gdc-data-transfer-tool). After downloading the count data which is presented as the Roshal Archive, the gene expression matrix with ensemble ID was extracted by decompressing packages and extracting the script manually. Downloading of the TCGA symbol IDs for sampling was used to correlate gene expression matrices using R (version: 2.15.3). These samples were classified into two groups: a tumour group (n=1,102) and a normal group (n=113). Gene expression data were compared between the two groups by Student’s t-test. Approval by an ethics committee was not needed as the data were obtained from TCGA. This study meets the TCGA publication guidelines.
Patients and tumor specimens
The study was approved by the Medical Ethics Committee of Xinxiang Central Hospital. Paraffin-embedded tissue blocks from 96 IDC patients who underwent biopsy at initial diagnosis between April 2001 and August 2004 without treatment at initial presentation were retrieved. 67 normal breast tissues and 37 breast tissues of ductal carcinoma in situ (DCIS) were also included in the study. Three pathologists re-appraised the histological subtypes according to the current WHO classification. Tumor staging was re-appraised using the 7th edition of the American Joint Committee on Cancer (AJCC) system.
Survival analysis
All 96 IDC patients underwent chemotherapy and radiotherapy according to NCCN guidelines. The 37 patients with DCIS underwent a conventional surgical excision. Patient follow-up was defined as the time between surgery and the last hospital contact (scheduled follow-up or telephone contact) or a recurrence of the disease. The endpoints analyzed were disease-free survival (DFS) and overall survival (OS) and were calculated using the date of surgical resection to the date of the event. The event was defined as the time of hospital contact (scheduled follow-up or telephone contact), disease recurrence, or patient death according to the definition of DFS and OS. Patients lost to follow-up were recorded as the latest follow-up date. Ninety-six IDC tissues were divided into a high-transcription group and low-transcription group by the median value of FPKM of UHRF1 and analyzed by chi-square or Fisher’s exact test. The follow-up deadline was July 2014.
Immunohistochemical staining and assessment of UHRF1
Immunohistochemical staining was carried out as follows. Sections (4 µm thick) were cut from paraffin-embedded blocks and mounted on glass slides. The slides were deparaffinized with xylene and rehydrated with ethanol. The slides were heated in a pressure cooker with 10 mM citrate buffer solution (pH 6) for 2 minutes to retrieve antigen epitopes. Endogenous peroxidase was quenched by a 3% H2O2 treatment solution. The slides were washed with phosphate buffered saline (PBS) and incubated with blocking buffer at room temperature for 30 minutes, and then with UHRF1 primary antibodies (cat no: ab194236, 1:100, Abcam, UK) at 4 °C overnight followed by incubation with the secondary antibody at room temperature (Neobioscience, China) for 30 minutes. The slides were developed with 3,3-diaminobenzidine for 5 minutes and then counter-stained with hematoxylin. We used lymphocytes from human tonsils, which are known to express UHRF1, as the positive control and breast cancer tissue, incubated with PBS instead of the primary antibody, as the negative control.
Three pathologists, with no prior knowledge of the clinical and follow-up information, scored UHRF1 immunoexpression using a multi-head microscope to reach a consensus. Nuclear UHRF1 staining scores were calculated using H-scores. The H-score was calculated using the following equation: H-score = ΣPi × (i +1), where i stands for the intensity score of the slides (which ranged from 0 to 3), and Pi stands for the percentage of stained cells at each intensity (which ranged from 0 to 100%). The H-score ranges from 0 to 4.0, where 0 indicates that 100% of cells were negative (0), and 4.0 indicates that 100% of the cells were strongly stained (3+). H-scores higher than the median value were construed as UHRF1 overexpression.
Statistical analysis
R (version: 2.15.3) was used to analyze the DEGs between the normal breast tissues and IDC tissues and to match symbol ID to the TCGA Ensembl ID. SPSS 19 software package (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. We compared the data between groups by t-test and evaluated the association between UHRF1 expression status and various clinicopathological parameters by chi-square or Fisher’s exact test (if the theoretical frequency was smaller than 5). For survival analysis, we performed log-rank tests to evaluate the prognostic differences between the groups and plotted the survival curves using the Kaplan-Meier method with GraphPad Prism 5.0 (GraphPad Software, San Diego, CA, USA). For all analyses, P<0.05 was considered statistically significant.
Results
UHRF1 is upregulated in IDC
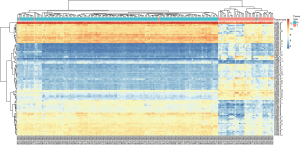
To identify the role of UHRF1 in the development and progression of breast cancer, the breast cancer datasets GSE10780 and GSE29044 were obtained from the GEO. In the GSE10780 dataset, we identified 4928 genes with adjusted P values <0.01, including 513 genes with log FC >1.5. Furthermore, the 513 DEGs identified consisted of 129 upregulated and 384 downregulated genes. UHRF1, TPX2, ADAMTS5, NUSAP1, COL10A1, ADAMTS5, DTL, UBE2T, CDK1, and NEK2 were the top 10 differentially expressed genes between IDC and normal tissues (Table 1). Among the statistically significant genes, UHRF1 had the smallest adjusted P value (1.79×10−55), and a log FC of 2.66, making it the most differentially expressed gene. We plotted the heat map of the top 100 differently expressed genes (Figure 1). In GSE29044, UHRF1 was also identified as a DEG with an adjusted P value of 2.07×10−6 and a log FC of 2.03.
Table 1
| Probe | Gene symbol | LogFC | Adj P value | Molecular function |
|---|---|---|---|---|
| 225655_at | UHRF1 | 2.66 | 1.79×10−55 | Transferase activity, ubiquitin protein ligase activity, metal ion binding, hemi-methylated DNA-binding, hemi-methylated DNA-binding proximal promoter sequence-specific DNA binding, nucleosomal histone binding, methylated histone binding, methyl-cpg binding, identical protein binding, histone binding |
| 210052_s_at | TPX2 | 1.90 | 2.83×10−49 | ATP binding, GTP binding, importin-alpha family protein binding, protein binding, protein kinase binding |
| 229357_at | ADAMTS5 | −2.70 | 4.92×10−49 | Extracellular matrix binding, heparin binding, integrin binding, metalloendopeptidase activity, metallopeptidase activity, protein binding, zinc ion binding |
| 218039_at | NUSAP1 | 2.68 | 5.20×10−49 | DNA binding, RNA binding, microtubule binding, protein binding |
| 205941_s_at | COL10A1 | 3.12 | 9.98×10−49 | Metal ion binding, protein binding, extracellular matrix structural constituent, molecular_function |
| 235368_at | ADAMTS5 | −2.68 | 1.39×10−48 | Extracellular matrix binding, heparin binding, integrin binding, metalloendopeptidase activity, peptidase activity, metallopeptidase activity, protein binding, zinc ion binding |
| 218585_s_at | DTL | 2.26 | 1.39×10−48 | Protein binding, contributes_to ubiquitin-protein transferase activity |
| 223229_at | UBE2T | 2.32 | 1.45×10−48 | ATP binding, chromatin binding, protein binding, nucleotide binding, transferase activity, ubiquitin conjugating enzyme activity, ubiquitin protein ligase activity, ubiquitin protein ligase binding, ubiquitin-protein transferase activity |
| 203213_at | CDK1 | 2.46 | 1.90×10−48 | ATP binding, Hsp70 protein binding, RNA polymerase II carboxy-terminal domain kinase activity, RNA polymerase II carboxy-terminal domain kinase activity, chromatin binding, cyclin binding, cyclin-dependent protein serine/threonine kinase activity, RNA polymerase II carboxy-terminal domain kinase activity, protein serine/threonine kinase activity |
| 204641_at | NEK2 | 2.47 | 3.63×10−48 | ATP binding, metal ion binding, protein binding, protein kinase activity, protein phosphatase binding, protein serine/threonine kinase activity, transferase activity |
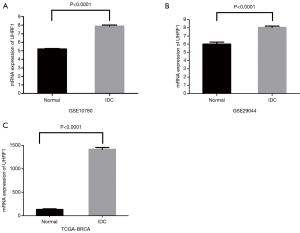
Compared with the normal breast tissues, expression of UHRF1 in GSE10780 and GSE29044 was significantly upregulated (Figure 2A,B respectively). We subsequently performed cross-validation using TCGA datasets and found that the UHRF1 expression in IDC was dramatically higher (10.65-fold) than that in paracarcinoma tissues (Figure 2C).
To further observe the expression of UHRF1 in breast cancer, immunohistochemistry was utilized to detect the expression of UHRF1 expression in 96 IDC samples, 37 DICS samples and 67 normal breast tissue samples. Positive staining of UHRF1 was mainly distributed in the nucleus of breast cancer. Representative staining of UHRF1 in IDC, DICS, and normal tissues are summarized in Figure 3A. In 96 cases of IDC, 60 (62.5%) showed UHRF1 overexpression. In 37 cases of DCIS, 20 showed UHRF1 overexpression (54.1%). In 67 cases of normal breast tissues, 21 showed UHRF1 overexpression (30.9%).
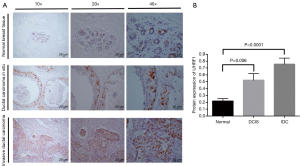
Subsequently, we compared the UHRF1 expression in the normal breast tissues and in IDC and DICS breast cancer samples. Our results showed that UHRF1 expression was the lowest in the normal breast tissues (average protein expression values =0.22). DCIS showed intermediate expression (average protein expression values =0.52), and IDC had the highest expression of UHRF1 (average protein expression values =0.76). Statistical analyses revealed a significant increase of UHRF1 in DCIS and IDC tissues compared to normal tissues (Figure 3B).
Relationship between UHRF1 and clinicopathological features of IDC
To dissect the roles of UHRF1 in the development of breast cancer, we analyzed the associations of UHRF1 expression with clinicopathological features of breast cancer using SPSS 19.0 software. We classified the breast cancer patients into UHRF1 high and low expression groups based on the median expression level of UHRF1.
As shown in Table 2, UHRF1 protein expression was correlated with estrogen receptor (ER) (χ2=4.200, P=0.040) and pathological grade (χ2=4.798, P=0.028) of breast tumor, but not related with other clinical features, such as age and sex (P>0.028).
Table 2
| Clinical Pathological Character | UHRF1 protein level* | χ2 | P value | |
|---|---|---|---|---|
| Low [49] | High [47] | |||
| Age | 1.841 | 0.175 | ||
| ≤60 | 29 | 34 | ||
| >60 | 20 | 13 | ||
| Location | 3.414 | 0.065 | ||
| Left | 28 | 18 | ||
| Right | 21 | 29 | ||
| ER | 4.200 | 0.040 | ||
| Negative | 14 | 23 | ||
| Positive | 35 | 24 | ||
| PR | 1.570 | 0.210 | ||
| Negative | 24 | 29 | ||
| Positive | 25 | 18 | ||
| HER-2 | 0.626 | 0.429 | ||
| Negative | 33 | 28 | ||
| Positive | 16 | 19 | ||
| T stage | 0.011 | 0.917 | ||
| T1 | 10 | 10 | ||
| T2–T3 | 39 | 37 | ||
| N stage# | 0.048 | 0.826 | ||
| N0–N1 | 32 | 31 | ||
| N2–N3 | 15 | 16 | ||
| Clinical stage# | 0.047 | 0.829 | ||
| I–II | 31 | 30 | ||
| III–IV | 16 | 17 | ||
| Pathological grade | 4.798 | 0.028 | ||
| I–II | 44 | 34 | ||
| III | 5 | 13 | ||
#, there are two missing values in the UHRF1 low expression group; *, 96 IDC tissues were divided into high-transcription group and low-transcription group by the median value of FPKM of UHRF1. UHRF1, ubiquitin-like PHD and RING finger domain-containing protein 1; IDC, invasive ductal carcinoma.
High UHRF1 expression is associated with poor survival in IDC
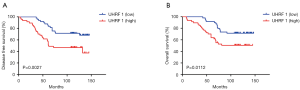
To explore the prognostic value of UHRF1 in breast cancer, Kaplan-Meier survival analysis was employed to analyze the DFS and OS. Among the total IDC patients, the average follow-up time was 97.3 (range, 8–147) months. As shown in Figure 4, patients with high UHRF1 expression had a worse OS (47.92%) than those with low UHRF1 expression (29.17%). The DFS of UHRF1 high expression and low expression groups were 31.25% and 54.17%, respectively, while the UHRF1 high expression group showed statistically significantly lower DFS (Figure 4A) and OS (Figure 4B) than that of the low expression group.
Discussion
Recent studies have demonstrated that UHRF1 is related to the progression (21), drug resistance (13), and radiotherapy (22) of breast cancer. Most of these studies were focused on dissecting the mechanism of how UHRF1 regulates breast cancer progression (23). However, very few studies have analyzed the correlation of the UHRF1 expression in breast cancer tissue by using large-scale patient population. In the current study, we investigated the role of UHRF1 in the development of breast cancer by integrating the GEO, TCGA datasets, and immunohistochemistry analysis. In this study, we analyzed not only the mRNA expression but also the protein expression of UHRF1 by multiple analyses, which can provide a comprehensive understanding of the role of UHRF1 in breast cancer.
In the GSE10780 and GSE29044 database, we observed that UHRF1 was significantly overexpressed in invasive ductal carcinoma tissue which is the most common pathological type of breast cancer (24). The data from the TCGA dataset showed that UHRF1 expression was increased in invasive ductal carcinoma compared to the normal breast tissue samples. Notably, we further explored the expression of UHRF1 in breast cancer tissues via immunohistochemistry to validate the results observed from the data garnered from the GEO and TCGA databases. The results demonstrated that UHRF1 was increased in breast cancer tissues, particularly in IDC. Furthermore, UHRF1 expression in DICS, which is the interim stage in the development of IDC was also analyzed. We found an increasing trend of UHRF1 expression across the DCIS and IDC tissues. However, there is no statistical significance between DCIS and IDC, and this might have been caused by insufficient specimens of DCIS. These integrated analyses indicated that UHRF1 is highly expressed and may be linked to the development of breast cancer.
ERs play an important role in breast cancer development. ER, progesterone receptor (PR), and human epidermal growth factor receptor-2 (HER2) are crucial biomarkers for predicting the response to hormone treatments for breast cancer (25). Compared to ER-positive tumors, triple-negative (ER-negative, PR negative, and Her-2 not overexpressed) breast cancers do not respond to some treatments and tend to be more aggressive (26). Macaluso et al. (27) found that the triple-negative breast cancer cell lines, MDA-MD-231 and MDA-MB-361, exhibited a higher expression of UHRF1 (ICBP90) protein levels than MCF-7 in western blotting. No study has investigated the correlation of UHRF1 expression in breast cancer tissue and ER expression by using large amounts of cancer tissues. In the present study, high UHRF1 levels in tissue were significantly associated with an ER-negative status. However, the mechanism by which UHRF1 regulates ER expression remains unclear. However, UHRF1 gene expression is closely related to the gene methylation levels (28). UHRF1 (ICBP90) can cooperate with pRb2/p130 and regulate the ER-α gene expression (27). Similar to previous findings, we also found that UHRF1 expression is highly correlated to the pathological grade of breast cancer (6).
High expression of UHRF1 is associated with poor prognosis in many cancers (10-12,21,29). In the survival analysis, we found that UHRF1 expression is distinctly related to DFS and OS in breast cancer patients. Similar results were reported in other cancers, including esophageal squamous cell carcinoma (30) and hepatocellular carcinoma (29). In breast cancer, elevated UHRF1 DNA levels in plasma were reported to be directly related with short, progression-free survival (20). The mechanism of UHRF1 leading to poor prognosis might be caused by be promotion of cell proliferation and metastasis since, in one published report, elevated UHRF1 expression contributed to poor prognosis by promoting cell proliferation and metastasis in hepatocellular carcinoma in a published report (10). Gao et al. (21) found that the overexpression of UHRF1 was linked to breast cancer progression and poor prognosis by suppression of KLF17 which plays a pivotal role in the epithelial-mesenchymal transition.
Conclusions
In conclusion, our study assessed the relationship between the expression of UHRF1 and the clinical outcomes of breast cancer patients by analyzing the data from the GEO and TCGA databases and validating those results via an immunohistochemistry analysis. Our results demonstrated that the levels of UHRF1 were increased in breast cancer and were associated with the ER expression and pathological tumor grade. More significantly, highly expressed UHRF1 predicted a poor prognosis. Although further studies are needed, our findings indicate that UHRF1 might promote tumorigenesis and the development of breast cancer.
Acknowledgments
Thanks for the help of Mrs. Lijun Chen and all members of Department of Pathology of Xinxiang Center Hospital.
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.06.19). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee of Xinxiang Central Hospital. Individual informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- DeSantis C, Ma J, Bryan L, et al. Breast cancer statistics, 2013. CA Cancer J Clin 2014;64:52-62. [Crossref] [PubMed]
- McDonnell DP, Park S, Goulet MT, et al. Obesity, cholesterol metabolism, and breast cancer pathogenesis. Cancer Res 2014;74:4976-82. [Crossref] [PubMed]
- Papavasiliou KA, Kirkos JM, Kapetanos GA, et al. Potential influence of hormones in the development of slipped capital femoral epiphysis: a preliminary study. J Pediatr Orthop B 2007;16:1-5. [Crossref] [PubMed]
- Hilakivi-Clarke L, Andrade JE, Helferich W. Is soy consumption good or bad for the breast? J Nutr 2010;140:2326S-2334S. [Crossref] [PubMed]
- Mousli M, Hopfner R, Abbady A, et al. ICBP90 belongs to a new family of proteins with an expression that is deregulated in cancer cells. Br J Cancer 2003;89:120. [Crossref] [PubMed]
- Jenkins Y, Markovtsov V, Lang W, et al. Critical role of the ubiquitin ligase activity of UHRF1, a nuclear RING finger protein, in tumor cell growth. Mol Biol Cell 2005;16:5621-9. [Crossref] [PubMed]
- Ferry L, Fournier A, Tsusaka T, et al. Methylation of DNA ligase 1 by G9a/GLP recruits UHRF1 to replicating DNA and regulates DNA methylation. Mol Cell 2017;67:550-565.e5. [Crossref] [PubMed]
- Patnaik D, Estève PO, Pradhan SJO. Targeting the SET and RING-associated (SRA) domain of ubiquitin-like, PHD and ring finger–containing 1 (UHRF1) for anti-cancer drug development. Oncotarget 2018;9:26243. [Crossref] [PubMed]
- Zhuo H, Tang J, Lin Z, et al. The aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinoma. Mol Carcinog 2016;55:209-19. [Crossref] [PubMed]
- Cui L, Chen J, Zhang Q, et al. Up-regulation of UHRF1 by oncogenic Ras promoted the growth, migration, and metastasis of pancreatic cancer cells. Mol Cell Biochem 2015;400:223-32. [Crossref] [PubMed]
- Unoki M, Kelly J, Neal D, et al. UHRF1 is a novel molecular marker for diagnosis and the prognosis of bladder cancer. Br J Cancer 2009;101:98. [Crossref] [PubMed]
- Jin W, Liu Y, Xu SG, et al. UHRF1 inhibits MDR1 gene transcription and sensitizes breast cancer cells to anticancer drugs. Breast Cancer Res Treat 2010;124:39-48. [Crossref] [PubMed]
- Jin W, Chen L, Chen Y, et al. UHRF1 is associated with epigenetic silencing of BRCA1 in sporadic breast cancer. Breast Cancer Res Treat 2010;123:359-73. [Crossref] [PubMed]
- Zhang Q, Qiao L, Wang X, et al. UHRF1 epigenetically down-regulates UbcH8 to inhibit apoptosis in cervical cancer cells. Cell Cycle 2018;17:300-8. [Crossref] [PubMed]
- Geng Y, Gao Y, Ju H, et al. Diagnostic and prognostic value of plasma and tissue ubiquitin-like, containing PHD and RING finger domains 1 in breast cancer patients. Cancer Sci 2013;104:194-9. [Crossref] [PubMed]
- Unoki M, Nishidate T, Nakamura YJO. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene 2004;23:7601. [Crossref] [PubMed]
- Adachi Y, Ishiguro J, Kotani H, et al. Comparison of clinical outcomes between luminal invasive ductal carcinoma and luminal invasive lobular carcinoma. BMC Cancer 2016;16:248. [Crossref] [PubMed]
- Chen DT, Nasir A, Culhane A, et al. Proliferative genes dominate malignancy-risk gene signature in histologically-normal breast tissue. Breast Cancer Res Treat 2010;119:335-46. [Crossref] [PubMed]
- Colak D, Nofal A, AlBakheet A, et al. Age-specific gene expression signatures for breast tumors and cross-species conserved potential cancer progression markers in young women. PLoS One 2013;8:e63204. [Crossref] [PubMed]
- Gao SP, Sun HF, Li LD, et al. UHRF1 promotes breast cancer progression by suppressing KLF17 expression by hypermethylating its promoter. Am J Cancer Res 2017;7:1554. [PubMed]
- Li X, Meng Q, Rosen EM, et al. UHRF1 confers radioresistance to human breast cancer cells. Int J Radiat Biol 2011;87:263-73. [Crossref] [PubMed]
- Li XL, Xu JH, Nie JH, et al. Exogenous expression of UHRF1 promotes proliferation and metastasis of breast cancer cells. Oncol Rep 2012;28:375-83. [PubMed]
- Guiu S, Wolfer A, Jacot W, et al. Invasive lobular breast cancer and its variants: how special are they for systemic therapy decisions? Crit Rev Oncol Hematol 2014;92:235-57. [Crossref] [PubMed]
- Parise CA, Bauer KR, Brown MM, et al. Breast cancer subtypes as defined by the estrogen receptor (ER), progesterone receptor (PR), and the human epidermal growth factor receptor 2 (HER2) among women with invasive breast cancer in California, 1999–2004. Breast J 2009;15:593-602. [Crossref] [PubMed]
- Brouckaert O, Wildiers H, Floris G, et al. Update on triple-negative breast cancer: prognosis and management strategies. Int J Womens Health 2012;4:511. [PubMed]
- Macaluso M, Montanari M, Noto PB, et al. Epigenetic modulation of estrogen receptor-α by pRb family proteins: a novel mechanism in breast cancer. Cancer Res 2007;67:7731-7. [Crossref] [PubMed]
- Rothbart SB, Krajewski K, Nady N, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol 2012;19:1155. [Crossref] [PubMed]
- Liang D, Xue H, Yu Y, et al. Elevated expression of UHRF1 predicts unfavorable prognosis for patients with hepatocellular carcinoma. Int J Clin Exp Pathol 2015;8:9416. [PubMed]
- Ye J, Zhang Y, Liang W, et al. UHRF1 is an independent prognostic factor and a potential therapeutic target of esophageal squamous cell carcinoma. J Cancer 2017;8:4027. [Crossref] [PubMed]


