
Five tumor-associated autoantibodies expression levels in serum predict lung cancer and associate with poor outcome
Introduction
Lung cancer is the most common cause of death from malignant cancer worldwide (1,2). Surgical resection is the primary and effective option for lung cancer, but only 20% to 25% of patients are eligible for the resection (3). The majority of patients with lung cancer are diagnosed with advanced disease owing to the paucity of early diagnostic methods. Biopsy and pathologic examination were widely used to diagnose lung cancer when lesion was discovered by chest X-ray or computerized tomography (CT). A number of biomarkers such as carcinoembryonic antigen (CEA) in blood have been identified in the clinic to assist diagnosis of lung cancer (4,5). Due to low expression of these biomarkers, they had limited sensitivity to be considered as appropriately screening method for lung cancer (6,7).
Autoantibody which can reflect a hosts’ immune response to tumor-associated antigens (TAAs) have been validated for early detection of lung cancer and subsequent treatment (8-10). Currently, we aimed to explored the autoantibodies for screening lung cancer. We extracted TAAs for lung cancer from the cancer genome atlas (TCGA) database and further describe how expression status of the corresponding tumor-associated autoantibodies in serum to assess the malignance of pulmonary nodules and their significant in the clinic.
Methods
Patients and specimens
In current study, 352 sera were consecutively collected from the patients with primary lung cancer which were diagnosed by pathological examination through thoracotomy or needle biopsy and 69 of tumor tissues were also got among these patients from January 2015 to January 2017 in the Department of Thoracic Surgery II of Peking University Cancer Hospital. Those patients with lung benign lesion were excluded. In the meantime, 226 cases normal individuals with healthy physical examination were included in this study from the Department of Thoracic Surgery of Jilin Guowen Hospital. Those individuals with malignant tumor history were excluded. The stage was classified according to tumor-lymph node-metastasis (TMN) classification system and the American Joint Committee on Cancer (AJCC). Three hundred and forty-nine cases of patients were detected with carcino-embryonic antigen (CEA) in our department of clinical laboratory. Two hundred and eighty-six patients were examined by positron emission computed tomography (PET)-CT. In addition, 226 cases of sera from normal individuals with healthy physical examination. The participants signed informed consent and this study has been approved with the Ethics Committee of Peking University Cancer Hospital.
Immunohistochemical staining and score evaluation
According to our previous study (11), immunohistochemistry assay was done. Briefly, formalin-fixed sections were rehydrated using the graded serious ethanol solution. Antigen retrieval was performed in the 10 mM citrate buffer at 98–100 °C for 30 min and 10% normal goat serum was used to block the unspecific binding for 30 min. The primary antibodies including mouse monoclonal anti-GAGE7 antibody (1:100 dilution; Abcam, UK), mouse monoclonal anti-MAGEA1 antibody (1:100 dilution; Absin, China), mouse monoclonal anti-PGP9.5 antibody (1:200 dilution; Abcam, UK), mouse monoclonal anti-CAGE antibody (1:100 dilution; Absin, China) and mouse monoclonal anti-p53 antibody (1:200 dilution; Abcam, UK) were added on the section overnight at 4 °C, followed by incubation with HRP-conjugated goat anti-mouse secondary antibody for 30 min at room temperature (Sigma-Aldrich, Poole, Dorset, UK).
Immunohistochemical evaluation was graded based on the intensity of staining (1+, weak; 2+, moderate; 3+, strong) and the percentage of stained cells (0, for <5%; 1, for 5–25%; 2, for 26–50% and 3, >50%) (12). The score used for the analyses was the average values. When combining these two parameters, 0–1 and >1 were considered negative and positive staining, respectively. Scoring was reviewed in parallel by two experienced pathologists who were blinded to all clinical data.
Enzyme-linked immunosorbent assay (ELISA)
Serum samples from patients and healthy individuals were tested by ELISA assay. Purified recombinant proteins of GAGE7, MAGEA1, PGP9.5 CAGE and p53, were coated onto 96-well microtiter plates (CancerProbe, China). Serum samples from patients and healthy individuals were diluted at 1:100 and added in the plated for 1h at room temperature. After the samples were washed with PBS, HRP-conjugated secondary antibody was used to incubating the samples for 30 min. The optical density value of each well was read at 450 nm, and the relative expression levels of the five autoantibodies were calculated based on the standard curve of each plate. The value for concentration of the 5 autoantibodies in serum were defined as unit (U). This experiment for all the samples were tested in duplicate.
Statistical analysis
Pearson’s correlation method was used to analyze the correlation of autoantibodies expression between serum and lung cancer tissues. Associations between the autoantibodies’ expressions and patients’ clinical variables were assessed using t-test. The receiver operating characteristic (ROC) analysis and logistic regression were used to evaluate the diagnostic value of 5 autoantibodies. When we constructed the logistic model to evaluate the combination of five autoantibodies, we used the multivariate regression analysis to calculate the prediction probability of combination of five autoantibodies for each patient and then used the ROC analysis to evaluate the diagnostic value. All statistical analyses were performed using SPSS 23.0 statistical software. The data are presented as the means ± standard error of the mean (SEM). P<0.05 was considered significant difference.
Results
Screening of TAAs for lung cancer patients
Firstly, we selected the commonly reported TAAs including AnnexinA1 (13,14), AnnexinA2 (15), CathepsinD (16), CyclinD1 (17), p53 (18), PGP9.5 (19), Recoverin (20), Paxillin (21), MALAT1 (22), GAGE7 (23), CAGE (24,25), MAGEA1 (26) and Myc (27) and analyzed these autoantibodies expressions in the 403 cases of lung cancers (241 cases of adenocarcinoma and 162 squamous cell carcinoma) and 57 cases of normal lung tissues from TCGA database of RNA-sequence (RNA-SEQ) (Figure 1A). GAGE7, MAGEA1, PGP9.5, CAGE and p53 were observed to be highly expressed in the lung cancer tissues compared to normal tissues with more than 1.5-fold increase.
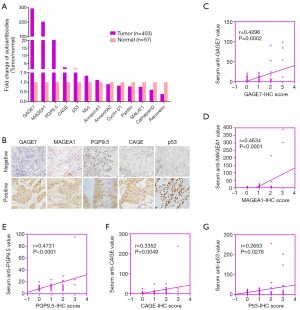
Correlation of tumor-associated autoantibodies between tumor tissues and serum of lung cancer patients
Then, we detected GAGE7, MAGEA1, PGP9.5, CAGE and p53 protein expression in 69 cases of lung cancer tissues by immunohistochemistry (Figure 1B). Moreover, these five autoantibodies were also investigated in the corresponding serum samples from these 69 lung cancer patients. As shown in Figure 1C,D,E,F,G and Table 1, positive association were shown in lung cancer tissues and serum samples for GAGE7, MAGEA1, PGP9.5, CAGE and p53 detection (r=0.4296, P=0.0002; r=0.4634, P<0.0001; r=0.4731, P<0.0001; r=0.3352, P=0.0049; r=0.2653, P=0.0276; respectively). The results indicated that expression level of each single autoantibody in serum were consistent with their protein expression degrees in lung cancer tissues.
Table 1
| Autoantibodies | Negative, N (%) | 1+, N (%) | 2+, N (%) | 3+, N (%) | r value | P value |
|---|---|---|---|---|---|---|
| GAGE7 | 17 (24.6) | 30 (43.5) | 18 (26.1) | 4 (5.8) | 0.4296 | 0.0002 |
| MAGEA1 | 11 (15.9) | 37 (53.6) | 19 (27.5) | 2 (2.9) | 0.4634 | <0.0001 |
| PGP9.5 | 38 (55.1) | 17 (24.6) | 11 (15.9) | 3 (4.3) | 0.4731 | <0.0001 |
| CAGE | 27 (39.1) | 17 (24.6) | 22 (31.9) | 3 (4.3) | 0.3352 | 0.0049 |
| p53 | 25 (36.2) | 18 (26.1) | 11 (15.9) | 15 (21.7) | 0.2653 | 0.0276 |
Diagnostic potential of tumor-associated autoantibodies in serum for lung cancer
We asked how expression levels of the autoantibodies such as GAGE7, MAGEA1, PGP9.5, CAGE and p53 varied in serum between the patients with lung cancer and normal individuals. We tested the concentration of these five autoantibodies in serum from 352 lung cancer patients and 226 normal individuals. For GAGE7, MAGEA1, PGP9.5, CAGE and p53 expression, there were 25.3±4.6, 12.8±2.3, 14.8±2.2, 11.7±1.5, 19.6±2.2 U/mL in lung cancer patients and 3.8±0.2, 5.0±1.5, 5.0±0.3, 4.6±0.7, 8.2±1.4 U/mL in normal individuals, respectively (Figure 2A). A t-test showed, there were significantly higher levels of these five autoantibodies in lung cancer patients compared to the normal individuals (P<0.0001, 0.005, <0.0001, <0.0001, <0.0001 for GAGE7, MAGEA1, PGP9.5, CAGE and p53, respectively).
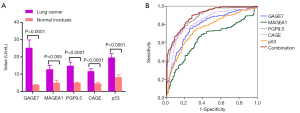
We further analyzed whether these five autoantibodies could discriminate the patients with lung cancer and normal individuals. The sensitivity, specificity, optimal cut-off value and area under curve (AUC) of each autoantibody were demonstrated in Table 2. To estimate the risk of individuals being diagnosed with lung cancer, ecoefficiency value was generated for each autoantibody by logistic regression model (Table 2). The combinational predictive probability was then created as following Eq.: PRE = 1/[1 + EXP (−3.054 + 0.103 × GAGE7 + 0.001 × MAGEA1 + 0.406 × PGP9.5 + 0.006 × CAGE + 0.002 × p53)]. For combinational autoantibodies, we could observe 90.1%, 71.7% and 0.891 for the sensitivity, specificity and AUC (Table 2), showing better predictive performance than each signal autoantibody (Figure 2B).
Table 2
| Autoantibodies | Sensitivity | Specificity | Cut-off value (U/mL) | AUC (95% CI) | Coefficiency |
|---|---|---|---|---|---|
| GAGE7 | 0.756 | 0.739 | 3.7 | 0.787 (0.749–0.825) | 0.103 |
| MAGEA1 | 0.364 | 0.85 | 3.4 | 0.641 (0.596–0.686) | 0.001 |
| PGP9.5 | 0.818 | 0.757 | 6.2 | 0.873 (0.844–0.902) | 0.406 |
| CAGE | 0.739 | 0.832 | 3.8 | 0.825 (0.789–0.861) | 0.006 |
| p53 | 0.724 | 0.704 | 6.1 | 0.749 (0.708–0.789) | 0.002 |
| Combination | 0.901 | 0.717 | 0.5008* | 0.891 (0.864–0.917) | – |
*, predictive probability. ROC, receiver operating characteristic; AUC, area under curve.
Clinical significance of five tumor-associated autoantibodies for lung cancer patients
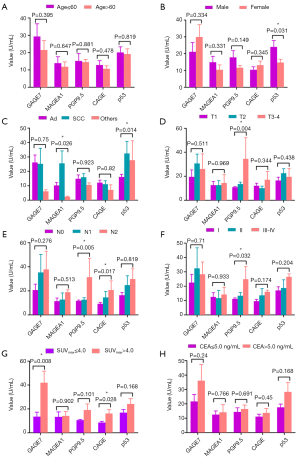
We further analyzed relationship between expression status of GAGE7, MAGEA1, PGP9.5, CAGE and p53 in serum from 352 lung cancer patients and their clinical characteristics like age, gender, pathological type, T stage, N stage and TNM stage, SUVmax of primary tumor and CEA level. The clinical features for these patients are summarized in Table 3. There was no statistically significant difference between age and any autoantibody (Figure 3A). The p53 level was higher in male than female (Figure 3B, P=0.031). The level of MAGEA1 and p53 were highly expressed in patients of squamous cell carcinoma than adenocarcinoma and other pathological types (Figure 3C, P=0.026 and 0.014, respectively). Furthermore, increased levels for each autoantibody was found, when the T stage, N stage and TNM stage got more advanced (Figure 3D,E,F). Among these five autoantibodies, PGP9.5 level was significantly elevated in serious T stage, N stage and TNM stage (P=0.004, 0.005, and 0.032, respectively). The CAGE was significantly higher in serious N stage (P=0.017). Increase of every autoantibody concentration in serum was demonstrated in the lung cancer patients with SUVmax of tumor >4.0 than SUVmax ≤4.0, and there were statistically significant differences for GAGE7 and CAGE (Figure 3G, P=0.008 and 0.028, respectively). The expression level for any autoantibody was higher in the patients with positive CEA (CEA >5.0 ng/mL) than the patients with negative CEA, although the difference was not statistically significant (Figure 3H). These results indicated that these autoantibodies were associated with gender, pathological type, T stage, N stage and TNM stage and SUVmax of primary tumor for the lung cancer patients.
Table 3
| Variables | Lung cancers, N (%) |
|---|---|
| Age | |
| ≤60 | 168 (47.7) |
| >60 | 184 (52.3) |
| Gender | |
| Male | 184 (52.3) |
| Female | 168 (47.7) |
| Smoking history | |
| Yes | 143 (40.6) |
| No | 209 (59.4) |
| Drinking history | |
| Yes | 75 (21.3) |
| No | 277 (78.7) |
| Pathological type | |
| Adenocarcinoma | 275 (78.1) |
| Squamous cell carcinoma | 66 (18.8) |
| Others | 11 (3.1) |
| CEA (ng/mL) | |
| ≤5.0 | 259 (74.2) |
| >5.0 | 90 (25.8) |
| Tumor size (cm) | |
| ≤3.0 | 240 (68.2) |
| >3.0 | 112 (31.8) |
| SUVmax | |
| ≤4.0 | 134 (38.1) |
| >4.0 | 152 (43.2) |
| Unknown | 66 (18.8) |
| EGFR status | |
| Wild | 57 (16.2) |
| Mutation | 136 (38.6) |
| Unknown | 193 (54.8) |
| T stage | |
| T1 | 152 (43.2) |
| T2 | 159 (45.2) |
| T3–4 | 41 (11.6) |
| N stage | |
| N0 | 249 (70.7) |
| N1 | 48 (13.6) |
| N2 | 55 (15.6) |
| TNM stage | |
| I | 216 (61.4) |
| II | 50 (14.2) |
| III–IV | 86 (24.4) |
CEA, carcinoembryonic antigen; EGFR, epidermal growth factor receptor.
Discussion
For lung cancer, diagnosis often occurs so late that approximately two-thirds of patients have lost the opportunity for radical surgery (28,29). Lung cancer screening using low-dose CT is recommended in select high-risk smokers, and there are 24.2% positive screening tests. It is necessary to identify the malignant lung nodules from positive screening results. Detection of autoantibody in serum to be potential biomarker for diagnosis of cancer has been developed (30,31).
With analysis of the TCGA database, we selected the highly expressed TAAs including GAGE7, MAGEA1, PGP9.5, CAGE and p53 genes in lung cancer tissues at mRNA level. Autoantibodies against GAGE7, MAGEA1 and CAGE which are members of the cancer testis antigen family, have also been reported in lung cancer (10,32). Antibodies recognizing p53 protein were first detected in breast cancer patients (33), and were later reported in the early stages of lung cancer as well (10). The antigen of PGP9.5 was a tumor suppressor gene that is inactivated by promoter methylation or gene deletion in several types of human cancers (34). In addition, PGP9.5 was previously identified as an antigen that induces an antibody response in lung cancer (35,36). We further analyzed the correlation between the expression of five autoantibodies against these TAAs in tumor tissues and the serum from lung cancer patients and found that the relationship was statistically significant. It was proved that autoantibodies against GAGE7, MAGEA1, PGP9.5, CAGE and p53 in serum could reflect the expression level of these antigens in primary tumors.
To discriminate lung cancer and healthy individuals, we firstly found that significantly higher level of GAGE7, MAGEA1, PGP9.5, CAGE and p53 in lung cancer patient than in individuals. Next, we could see high AUC value of 0.787, 0.641, 0.873, 0.825 and 0.749 for GAGE7, MAGEA1, PGP9.5, CAGE and p53 in ROC analysis, respectively. Previous studies have shown that single or combinations of multiplex autoantibodies can be used as a tool for the diagnosis of lung cancer patients at all stages or early-stage, and the combination of multiplex autoantibodies shows a higher detection capacity (37). Jia et al. also found multiply autoantibodies were more accurate to predict lung cancer than single autoantibody (38). With the encouraging results, we provided a hypothesis that whether a panel of these five autoantibodies could help diagnose lung cancer. ROC analysis indicated the accuracy of diagnosis was up to exciting 89.1% with sensitivity 90.1% and specificity 71.7% when combining the five antibodies. Compared to the CEA with 25.8% sensitivity in all 352 lung cancer patients of current study, five autoantibodies are suggested to be favorable biomarkers for detection of lung cancer. Maybe the five antibodies can be used to monitor the occurrence of lung cancer at an early stage by using a test kit to be developed.
Murray et al. proved acceptable concentration level of autoantibodies including p53, SOX2, NY-ESO-1, CAGE, GBU4-5, Annexin1 in serum that could identify 40% of primary lung cancer (9). Chapman et al. used ELISA assay to display significance of autoantibodies such as p53, c-Myc, HER2, NY-ESO-1, CAGE, MUC1 and GBU4-5 in diagnosis and prognosis for lung cancer patients (10). Currently, we selected higher levels tumor-associated autoantibodies of GAGE7, MAGEA1, PGP9.5, CAGE and p53 in lung cancer tissues from TCGA database. We also discovered the same autoantibodies of p53 and CAGE with the above reports in lung cancer. Furthermore, we clarify correlation between the expression of the autoantibody levels in serum and clinical characteristics for the 352 lung cancer patients. It was found that there was higher expression of each single autoantibody in CEA positive, SUVmax >4.0, advanced T stage and N stage group compared to CEA negative, SUVmax ≤4.0, early T stage and N stage, although the differences were not all significant. Especially, the PGP9.5 level was positively correlated with T stage, N stage and TNM stage with significant differences. Additionally, MAGEA1 and p53 highly expressed in the patients with pathological type of squamous cell carcinoma and increased level of GAGE7 and CAGE were observed in the high level of SUVmax for lung cancer patients with PET-CT examination. All in all, the autoantibody may reflect the prognosis of lung cancer and it is necessary to design further study.
Conclusions
In sum, our data demonstrate that the serum antibody levels of GAGE7, MAGEA1, PGP9.5, CAGE and p53 were apparently higher in lung cancer patients than normal individuals and in view of that they could be used as early diagnosing lung cancer with satisfactory sensitivity and accuracy. Therefore, detection kits for early diagnosis of lung cancer may be developed in the future. In addition, the serum levels of autoantibodies were correlated with the stage of lung cancer and the prognosis value would be evaluated in further study.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.07.25). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study has been carried out in accordance with the Declaration of Helsinki (as revised in 2013) and has been approved with the Ethics Committee of Peking University Cancer Hospital and the number of ethics approval is 2015KT71. The participants signed informed consent prior to sample collection.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013;63:11-30. [Crossref] [PubMed]
- Xue C, Hu Z, Jiang W, et al. National survey of the medical treatment status for non-small cell lung cancer (NSCLC) in China. Lung Cancer 2012;77:371-5. [Crossref] [PubMed]
- Molina R, Filella X, Augé JM, et al. Tumor markers (CEA, CA 125, CYFRA 21-1, SCC and NSE) in patients with non-small cell lung cancer as an aid in histological diagnosis and prognosis. Comparison with the main clinical and pathological prognostic factors. Tumour Biol 2003;24:209-18. [Crossref] [PubMed]
- Cedrés S, Nuñez I, Longo M, et al. Serum tumor markers CEA, CYFRA21-1, and CA-125 are associated with worse prognosis in advanced non-small-cell lung cancer (NSCLC). Clin Lung Cancer 2011;12:172-9. [Crossref] [PubMed]
- Ando S, Kimura H, Iwai N, et al. Optimal combination of seven tumour markers in prediction of advanced stage at first examination of patients with non-small cell lung cancer. Anticancer Res 2001;21:3085-92. [PubMed]
- Chen X, Wang X, He H, et al. Combination of circulating tumor cells with serum carcinoembryonic antigen enhances clinical prediction of non-small cell lung cancer. PLoS One 2015;10:e0126276. [Crossref] [PubMed]
- Boyle P, Chapman CJ, Holdenrieder S, et al. Clinical validation of an autoantibody test for lung cancer. Ann Oncol 2011;22:383-9. [Crossref] [PubMed]
- Murray A, Chapman CJ, Healey G, et al. Technical validation of an autoantibody test for lung cancer. Ann Oncol 2010;21:1687-93. [Crossref] [PubMed]
- Chapman CJ, Murray A, McElveen JE, et al. Autoantibodies in lung cancer: possibilities for early detection and subsequent cure. Thorax 2008;63:228-33. [Crossref] [PubMed]
- Ma Y, Li M, Si J, et al. Blockade of Notch3 inhibits the stem-like property and is associated with ALDH1A1 and CD44 via autophagy in non-small lung cancer. Int J Oncol 2016;48:2349-58. [Crossref] [PubMed]
- Tsaur I, Makarević J, Juengel E, et al. Resistance to the mTOR-inhibitor RAD001 elevates integrin α2- and β1-triggered motility, migration and invasion of prostate cancer cells. Br J Cancer 2012;107:847-55. [Crossref] [PubMed]
- Biaoxue R, Xiguang C, Hua L, et al. Increased level of annexin A1 in bronchoalveolar lavage fluid as a potential diagnostic indicator for lung cancer. Int J Biol Markers 2017;32:e132-e140. [Crossref] [PubMed]
- Rong B, Zhao C, Liu H, et al. Elevated serum annexin A1 as potential diagnostic marker for lung cancer: a retrospective case-control study. Am J Transl Res 2014;6:558-69. [PubMed]
- Yang J, Yang F, Nie J, et al. Evaluation of Annexin A2 as a novel diagnostic serum biomarker for lung cancer. Cancer Biomark 2015;15:205-11. [Crossref] [PubMed]
- Lou X, Xiao T, Zhao K, et al. Cathepsin D is secreted from M-BE cells: its potential role as a biomarker of lung cancer. J Proteome Res 2007;6:1083-92. [Crossref] [PubMed]
- Zhou C, An H, Hu M, et al. The cyclin D1 (CCND1) G870A polymorphism and lung cancer susceptibility: a meta-analysis. Tumour Biol 2013;34:3831-7. [Crossref] [PubMed]
- Mattioni M, Soddu S, Prodosmo A, et al. Prognostic role of serum p53 antibodies in lung cancer. BMC Cancer 2015;15:148. [Crossref] [PubMed]
- Sasaki H, Yukiue H, Moriyama S, et al. Expression of the protein gene product 9.5, PGP9.5, is correlated with T-status in non-small cell lung cancer. Jpn J Clin Oncol 2001;31:532-5. [Crossref] [PubMed]
- Drlicek M, Unterberger U. Serum antibodies against recoverin and Hu: two paraneoplastic epiphenomena in lung cancer. Lung Cancer 2005;47:143-4; author reply 145-6. [Crossref] [PubMed]
- Jagadeeswaran R, Surawska H, Krishnaswamy S, et al. Paxillin is a target for somatic mutations in lung cancer: implications for cell growth and invasion. Cancer Res 2008;68:132-42. [Crossref] [PubMed]
- Weber DG, Johnen G, Casjens S, et al. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res Notes 2013;6:518. [Crossref] [PubMed]
- Kao CS, Badve SS, Ulbright TM. The utility of immunostaining for NUT, GAGE7 and NY-ESO-1 in the diagnosis of spermatocytic seminoma. Histopathology 2014;65:35-44. [Crossref] [PubMed]
- Park S, Lim Y, Lee D, et al. Identification and characterization of a novel cancer/testis antigen gene CAGE-1. Biochim Biophys Acta 2003;1625:173-82. [Crossref] [PubMed]
- Por E, Byun HJ, Lee EJ, et al. The cancer/testis antigen CAGE with oncogenic potential stimulates cell proliferation by up-regulating cyclins D1 and E in an AP-1- and E2F-dependent manner. J Biol Chem 2010;285:14475-85. [Crossref] [PubMed]
- Cannuyer J, Loriot A, Parvizi GK, et al. Epigenetic hierarchy within the MAGEA1 cancer-germline gene: promoter DNA methylation dictates local histone modifications. PLoS One 2013;8:e58743. [Crossref] [PubMed]
- Li P, Shi JX, Dai LP, et al. Serum anti-MDM2 and anti-c-Myc autoantibodies as biomarkers in the early detection of lung cancer. Oncoimmunology 2016;5:e1138200. [Crossref] [PubMed]
- Hong QY, Wu GM, Qian GS, et al. Prevention and management of lung cancer in China. Cancer 2015;121:3080-8. [Crossref] [PubMed]
- She J, Yang P, Hong Q, et al. Lung cancer in China: challenges and interventions. Chest 2013;143:1117-26. [Crossref] [PubMed]
- Fan CW, Kuo YB, Lin GP, et al. Development of a multiplexed tumor-associated autoantibody-based blood test for the detection of colorectal cancer. Clin Chim Acta 2017;475:157-63. [Crossref] [PubMed]
- Desmetz C, Mange A, Maudelonde T, et al. Autoantibody signatures: progress and perspectives for early cancer detection. J Cell Mol Med 2011;15:2013-24. [Crossref] [PubMed]
- Zhou X, Yang F, Zhang T, et al. Heterogeneous expression of CT10, CT45 and GAGE7 antigens and their prognostic significance in human breast carcinoma. Jpn J Clin Oncol 2013;43:243-50. [Crossref] [PubMed]
- Crawford LV, Pim DC, Bulbrook RD. Detection of antibodies against the cellular protein p53 in sera from patients with breast cancer. Int J Cancer 1982;30:403-8. [Crossref] [PubMed]
- Tokumaru Y, Yamashita K, Kim MS, et al. The role of PGP9.5 as a tumor suppressor gene in human cancer. Int J Cancer 2008;123:753-9. [Crossref] [PubMed]
- Madoz-Gúrpide J, Kuick R, Wang H, et al. Integral protein microarrays for the identification of lung cancer antigens in sera that induce a humoral immune response. Mol Cell Proteomics 2008;7:268-81. [Crossref] [PubMed]
- Brichory F, Beer D, Le Naour F, et al. Proteomics-based identification of protein gene product 9.5 as a tumor antigen that induces a humoral immune response in lung cancer. Cancer Res 2001;61:7908-12. [PubMed]
- Tang ZM, Ling ZG, Wang CM, et al. Serum tumor-associated autoantibodies as diagnostic biomarkers for lung cancer: A systematic review and meta-analysis. PLoS One 2017;12:e0182117. [Crossref] [PubMed]
- Jia J, Wang W, Meng W, et al. Development of a multiplex autoantibody test for detection of lung cancer. PLoS One 2014;9:e95444. [Crossref] [PubMed]


