
Morphological and pathological features of basal-like breast cancer
Introduction
Breast cancer (BC) comprises a wide group of diseases characterized by different molecular subtypes with specific gene signatures and distinct clinical outcomes (1). Genomic studies, such as the PAM50 gene expression assay (2), allow to classify BC into at least five intrinsic subtypes comprising: luminal A (estrogen-receptor and/or progesterone-receptor positive, HER2 negative, and low expression of Ki-67) and luminal B (estrogen-receptor and/or progesterone-receptor positive, HER2 positive/negative with high levels of Ki-67) expressing luminal epithelial layer genes of the breast gland; “HER2-enriched” showing high expression/amplification of the HER2 receptor and adjacent genes on 17q12–21 chromosome; basal-like breast cancer (BLBC), with a pattern of expression similar to basal epithelial and normal myoepithelial cells of breast tissue; and “normal-like” BC, characterized by adipose and other non-epithelial genes expression and high basal-like and low luminal gene expression (2,3).
Approximately 15% of all BC are of BLBC subtype and represent a particularly aggressive tumor affecting mostly young women and associated with more aggressive behavior and worse prognosis (4). BLBCs are characterized by a high risk of brain and lung metastases, and by none correlations between the primary tumor size and regional lymph node metastases rate, differently from other BC subtypes (5).
Histologically, the majority of BLBCs are invasive ductal carcinomas of no special type (IDC-NST) type, with high histological grade, higher mitotic indices, and a rife lymphocytic infiltrate (6). BLBC comprise a heterogeneous group of BC tumors and only the use of specific immunohistochemical marker panels allowed the correct stratification of the different entities (7):
- BLBC with the lack of estrogen receptor (ER), progesterone receptor (PR), and HER2 expression also defined as ‘triple-negative’ (TNBC) immunophenotype;
- BLBC with the expression of one or more high-molecular-weight/basal cytokeratins (CK5/6, CK14, and CK17);
- BLBC with the lack of expression of ER and HER2 together with expression of CK5/6 and/or EGFR;
- BLBC associated to CK5/6 and/or EGFR expression, and the lack of ER, PR, and HER2 receptor.
Although BLBCs and TNBCs are often confused, these two terms are not synonymous. As a matter of the fact, most of the TNBCs are of basal-like phenotype, but not all BCs expressing ‘basal’ markers are TNBC. Likewise considering their molecular gene profiles, not all BLBC lack ER, PR and HER2 expression and not all TNBCs show basal-like phenotype markers. Molecularly, BLBC are considered more homogeneous than TNBC, but the terminologies continue to be misused (7).
This mini-review summarizes the main morphological, pathological and molecular features of BLBC we will focus the attention also on new biomarkers with higher sensitivity and/or specificity that could improve the performance of diagnosis and management of this tumor subtype.
Microscopic characteristics and histological subtypes of BLBC
BLBCs originate from the outer cell layer of the ductal and lobular frames of the breast gland, nearby the basal membrane. BLBC cells are prevalently myoepithelial with epithelial and smooth muscle features, and consequently they express smooth muscle markers and myofilament proteins (8).
General characteristics of BLBC are tumor size larger than 2 cm, histological grade of 3, and high mitotic rate (average 25 mitoses/10 HPFs). Tumor cells reveal vesicular chromatin pattern and prominent nucleoli. A central necrosis (in the middle of the tumor islands) has been observed in about 65% of cases and a geographic necrosis in roughly 40% of BLBCs (9). Moreover, a marked lymphoplasmacytic infiltration is more frequent in BLBC than in non-basal-like cancers (9).
BLBC involves almost all different histological types of BC, including invasive ductal carcinoma, invasive lobular carcinoma, mixed carcinoma, mucinous carcinoma, metaplastic carcinoma, papillary carcinoma, medullary carcinoma, tubular carcinoma, apocrine carcinoma, micropapillary carcinoma, signet ring cell carcinoma, pleomorphic carcinoma, cribriform carcinoma and more rarely neuroendocrine carcinoma and atypical medullary carcinoma (10).
Invasive ductal carcinoma of no special type (IDC-NST) represents the most common histological type of BLBC. IDC-NST accounts for about 82% of the tumors and is characterized by a worse clinical behavior and prognosis (11). IDC shows a ductal proliferation with stromal invasion, frequently associated to foci of ductal carcinoma in situ (DCIS). However, most IDCs do not represent specific histotype and are defined as IDC-NST (12). They are characterized by pleomorphic cells with many mitoses and prominent nucleoli, and the cells are organized to form diffuse sheets, cords, nests, or singly distributed cells frequently with a ductal differentiation as shown in Figure 1 (12). Medullary carcinoma account for about 10% of BLBCs and is characterized by large and pleomorphic cells, poorly differentiated with scanty stroma and prominent lymphoid infiltration (13) (Figure 1C). Medullary BC is a BLBC with a favorable outcome, whereas non-medullary BLBC generally has a poor prognosis. Several gene expression profiling studies showed that medullary BC is associated with a specific molecular signature reflecting a TH1-type immune response (13). Metaplastic carcinoma represents about 10% of all BLBCs and is characterized by spindle, chondroid, osseous and rhabdoid cells with squamous epithelium or mesenchymal differentiation (14) (Figure 1). Pleomorphic carcinoma is very rare and account for about 2% of all BLBCs. Cells with eosinophilic cytoplasm and hyperchromatic eccentric nucleus with prominent nucleolus are predominant in this subtype (15). BLBCs can also display areas of invasive lobular and tubular carcinoma (tubule-lobular carcinoma) (16) (Figure 1B).
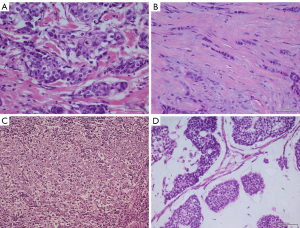
Basal-like mucinous carcinoma is of epithelial tumor cells with moderate nuclear atypia surrounded by abundant extracellular mucus (17) (Figure 1D). Basal-like neuroendocrine tumor cells are characterized by small, round or spindle cells, organized in alveolar, nest, trabecular, and rosette patterns (18).
Immunohistochemical markers
Typical immunohistochemical markers for BLBCs are represented by high basal-like cytokeratins (CK5/6, CK14, CK17), EGFR, P-cadherin, or c-kit expression and by lack of CD10, SMA, and p63 expression. Another distinguishing feature is the lack of expression of ER, PR, HER2 and high mitotic index or p53 aberrant expression (6).
About 50% of BLBCs are positive for both cytokeratin CK5/6 and EGFR, about 40% of them show positivity for CK5/6 and negativity for EGFR, whereas and a very small proportion of BLBCs is positive only for EGFR (6).
In about 27% of the BLBCs cytokeratin 14 is identified. Vimentin is expressed in about 52% of cases and both CK14 and vimentin co-expressions are significantly associated with the BLBC subtype (19).
Despite the molecular classification of basal-like tumors suggest a myoepithelial cell origin, most BLBC express luminal-type CKs, such as CK8/18, CK19, on the contrary only few cases express typical myoepithelial markers (such as actin and p63). About 50% of basal-like medullary carcinomas show strong immunoreactivity for P-cadherin, SMA and S100 (8).
BLBCs are characterized by higher expression of cell cycle genes such cyclin E1, BUB1, topoisomerase Iiα, CDC2, and PCNA and by Rb pathway inactivation (20). Moreover, fatty acid binding protein-7 (FABP-7) is significantly overexpressed in BLBCs and its co-expression with EGFR is strongly related with histological grade (21). About 50% of BLBCs shows the overexpression of matrix metalloproteinase-9 (MMP-9) and CD147 also correlated with poor prognosis and aggressive clinical course (22).
Caveolin 1 (CAV1) overexpression is associated with basal-like phenotype in both hereditary and sporadic BC. It is a marker of poorly differentiated BLBCs with poor prognosis (23).
Other emergent immunohistochemical biomarkers are represented by Mucin 1 (MUC1), a protein able to induce a specific immune response expressed in in 94% of BLBCs (24) and calretinin an intracellular, vitamin D-dependent calcium-binding protein, expressed in about 50% of BLBC especially in CK5/6 and EGFR-positive lesions and related with a very poor clinical outcome (25). Different cell surface molecules are overexpressed in BLBCs including nerve growth factor receptor (NGFR), CD44, CD280, c-Met and CD146, CD109 and placental cadherin (P-cadherin) (26). Several Extracellular Matrix (ECM) proteins are also aberrantly expressed in BLBCs, such as osteonectin and osteopontin (27). In particular, osteopontin has been found to be significantly higher in BLBCs and correlates with poor prognostic factors (28). Other ECM glycoproteins involved in cell adhesion mechanisms, such as laminins, are associated with the basal-like phenotype. In particular, the β4 integrin subunit is preferentially expressed in BLBC compared to non-basal-like cancer (29).
Finally, epithelial-mesenchymal transition (EMT) seems to play a key role in BLBC progression. Several EMT markers show a different expression in this tumor type: N-cadherin and vimentin are frequently overexpressed while E-cadherin is often lost (30).
Main molecular alterations
BLBCs are characterized by homogeneous molecular profile compared to all TNBCs. The most important molecular alteration is the heritable BRCA1 mutation. However, the most of BLBCs have a normal nuclear expression of BRCA1 (31), suggesting that the contribute of other genetic or epigenetic alterations in BRCA1-associated proteins might underlie the BRCA1 dysfunction phenotype of BLBC (32,33).
TP53 mutations have a high frequency (44–82%) in BLBC, which is responsible for the consequent interference with DNA repair mechanisms and apoptosis, consistently increasing the genetic instability (2,34,35). The loss of one TP53 allele in BRCA1 deleted animal model strongly promotes BC carcinogenesis, suggesting that p53 mutation synergistically act with BRCA1 defects during tumor evolution of BLBC (36).
EGFR is expressed in 39–54% of BLBC in which it promotes cell proliferation through Ras/MAPK/MAPK kinase pathway and confers resistance to apoptosis by ligand-dependent activation of the PI3-kinase/Akt/mTOR pathway (37-39). BLBC is also characterized by differential expression of cell cycle genes. RB and Cyclin D1 genes have a low expression whilst E2F3 and Cyclin E genes are abundantly expressed (40). In particular, Cyclin E1 is overexpressed in BLBC than other molecular subtypes of BC, and its expression is strongly associated with poor prognosis (41-43). Another common molecular alteration in BLBC is represented by the inactivation of the tumor suppressor gene PTEN able to lead the anomalous activation of the PI3-kinase/Akt/mTOR pathway (44-46). The loss of PTEN has been also associated with alterations in Rad51-mediated DNA double-strand break repair, able to promote genome instability in BLBC (47). Aberrant expression of molecular chaperone αB-crystallin in about 45% of BLBC leads the suppression of apoptosis by inhibiting the protease caspase-3 (48,49). Moreover, αB-crystallin expression is correlated with pre-surgery chemotherapy resistance and poor prognosis in BLBC patients (50).
Regarding the contribution of EMT in BLBC evolution, the down-regulation of E-cadherin expression is related with the activation of TGF-β, Wnt, and Notch pathways in turn implicated in the promotion of FOXC2, Twist, Slug, Snail, and LBX1 transcription factors (51-53).
In the last years, aberrant activity of several non-coding RNA molecules, both long non-coding RNAs (lncRNA) and microRNAs (miR) has been also associated with BLBC pathogenesis. The lncRNAs HOTAIRM1 and FOXCUT are overexpressed in BLBC than in non-basal BC subtypes and associated with its aggressive phenotype (54,55). Moreover, the knockdown of FOXCUT in BLBC cell models is able to inhibit cell migration and proliferation (55). The lncRNA HOTAIR is overexpressed in the basal-like MCF-7-TNR cells, and its binding with enhancer of zeste homolog 2 (EZH2) form a molecular complex involved in the maintenance of the basal-like phenotype. HOTAIR is also aberrantly expressed in MDA-MB-157 cells in which it modulates the expression of basal-like genes and control cell proliferation (56).
The role of microRNAs in BC has been widely documented, both as diagnostic and prognostic markers and as circulating markers (57,58). A large number of miRNA are differentially expressed between luminal and BLBCs such as mir-17, 17*, 18a, 19a/b, 20a and 106a (59,60). This expression is influenced by DNA copy number (60) and is able to promote tumor progression by reducing PTEN expression (61), by inducing cell migration and metastasis (62), and by inhibiting tumor suppressor genes ZBTB4 (63) and Rb (64). Loss of mir-375 and let-7a is involved in EMT in BLBC cells (65). Lastly, long non-coding RNAs have been shown to play a role in drug resistance in BC and are now widely studied in therapeutic monitoring, being easily identifiable also as circulating markers (66).
Conclusions
BLBCs represent a distinctive BC molecular subtype characterized by the expression of basal epithelial genes. They have an aggressive clinical behavior characterized by early relapse and worse survival. Microscopic findings suggest there are many significant morphological differences between basal-like and non-basal-like breast carcinomas and several new markers can be included in the immunohistochemical panel for distinguishing BLBCs. Moreover, many molecular abnormalities have been associated with BLBCs pathogenesis and progression, including BRCA1 dysfunction, p53 mutations, up-regulation of EGFR and TGF-β, inactivation of PTEN and the aberrant expression of many lncRNAs and miRNAs. These last could represent new specific markers useful both in the BLBC diagnosis and prognosis.
Acknowledgments
Funding: This study was supported by
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Emanuela Esposito and Michelino De Laurentiis) for the focused issue “Rare Tumors of the Breast” published in Translational Cancer Research. This article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.06.50). The focused issue “Rare Tumors of the Breast” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jönsson G, Staaf J, Vallon-Christersson J, et al. Genomic subtypes of breast cancer identified by array-comparative genomic hybridization display distinct molecular and clinical characteristics. Biological subtypes of breast cancer: Prognostic and therapeutic implications. Breast Cancer Res 2010;12:R42. [Crossref] [PubMed]
- Sørlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001;98:10869-74. [Crossref] [PubMed]
- Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000;406:747-52. [Crossref] [PubMed]
- Bauer KR, Brown M, Cress RD, et al. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer 2007;109:1721-8. [Crossref] [PubMed]
- Liu N, Yang Z, Liu X, et al. Lymph node status in different molecular subtype of breast cancer: triple negative tumours are more likely lymph node negative. Oncotarget 2017;8:55534-43. [PubMed]
- Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol 2008;26:2568-81. [Crossref] [PubMed]
- Seal MD, Chia SK. What is the difference between triple-negative and basal breast cancers? Cancer J 2010;16:12-6. [Crossref] [PubMed]
- Da Silva L, Clarke C, Lakhani SR. Demystifying basal‐like breast carcinomas. J Clin Pathol 2007;60:1328-32. [Crossref] [PubMed]
- Livasy CA, Karaca G, Nanda R, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol 2006;19:264-71. [Crossref] [PubMed]
- Dieci MV, Orvieto E, Dominici M, et al. Rare Breast Cancer Subtypes: Histological, Molecular, and Clinical Peculiarities. Oncologist 2014;19:805-13. [Crossref] [PubMed]
- Fulford LG, Reis-Filho JS, Ryder K, et al. Basal-like grade III invasive ductal carcinoma of the breast: patterns of metastasis and long-term survival. Breast Cancer Res 2007;9:R4. [Crossref] [PubMed]
- Fulford LG, Easton DF, Reis-Filho JS, et al. Specific morphological features predictive for the basal phenotype in grade 3 invasive ductal carcinoma of breast. Histopathology 2006;49:22-34. [Crossref] [PubMed]
- Marginean F, Rakha EA, Ho BC, et al. Histological features of medullary carcinoma and prognosis in triple-negative basal-like carcinomas of the breast. Mod Pathol 2010;23:1357-63. [Crossref] [PubMed]
- Weigelt B, Kreike B, Reis-Filho JS. Metaplastic breast carcinomas are basal-like breast cancers: a genomic profiling analysis. Breast Cancer Res Treat 2009;117:273-80. [Crossref] [PubMed]
- Cakir A, Gonul II, Uluoglu O. A comprehensive morphological study for basal-like breast carcinomas with comparison to nonbasal-like carcinomas. Diagn Pathol 2012;7:145. [Crossref] [PubMed]
- Harbhajanka A, Lamzabi I, Singh RI, et al. Correlation of clinicopathologic parameters and immunohistochemical features of triple-negative invasive lobular carcinoma. Appl Immunohistochem Mol Morphol 2014;22:e18-26. [Crossref] [PubMed]
- Kuroda N, Fujishima N, Inoue K, et al. Basal-like carcinoma of the breast: further evidence of the possibility that most metaplastic carcinomas may be actually basal-like carcinomas. Med Mol Morphol 2008;41:117-20. [Crossref] [PubMed]
- Inno A, Bogina G, Turazza M, et al. Neuroendocrine Carcinoma of the Breast: Current Evidence and Future Perspectives. Update on Immunohistochemical Analysis in Breast Lesions. Oncologist 2016;21:28-32. [Crossref] [PubMed]
- Sousa B, Paredes J, Milanezi F, et al. P-cadherin, vimentin and CK14 for identification of basal-like phenotype in breast carcinomas: an immunohistochemical study. Histol Histopathol 2010;25:963-74. [PubMed]
- Toft DJ, Cryns VL. Minireview: Basal-Like Breast Cancer: From Molecular Profiles to Targeted Therapies. Mol Endocrinol 2011;25:199-211. [Crossref] [PubMed]
- Tang XY, Umemura S, Tsukamoto H, et al. Overexpression of fatty acid binding protein-7 correlates with basal-like subtype of breast cancer. Pathol Res Pract 2010;206:98-101. [Crossref] [PubMed]
- Liu Y, Xin T, Jiang QY, et al. CD147, MMP9 expression and clinical significance of basal-like breast cancer. Med Oncol 2013;30:366. [Crossref] [PubMed]
- Savage K, Lambros MB, Robertson D, et al. Caveolin 1 Is Overexpressed and Amplified in a Subset of Basal-like and Metaplastic Breast Carcinomas: A Morphologic, Ultrastructural, Immunohistochemical, and in situ Hybridization Analysis. Clin Cancer Res 2007;13:90-101. [Crossref] [PubMed]
- Siroy A, Abdul-Karim FW, Miedler J, et al. MUC1 is expressed at high frequency in early-stage basal-like triple-negative breast cancer. Hum Pathol 2013;44:2159-66. [Crossref] [PubMed]
- Taliano RJ, Lu S, Singh K, et al. Calretinin expression in high-grade invasive ductal carcinoma of the breast is associated with basal-like subtype and unfavorable prognosis. Hum Pathol 2013;44:2743-50. [Crossref] [PubMed]
- Won JR, Gao D, Chow C, et al. A survey of immunohistochemical biomarkers for basal-like breast cancer against a gene expression profile gold standard. Mod Pathol 2013;26:1438-50. [Crossref] [PubMed]
- Wang X, Chao L, Ma G, et al. Increased expression of osteopontin in patients with triple-negative breast cancer. Eur J Clin Invest 2008;38:438-46. [Crossref] [PubMed]
- Ortiz-Martínez F, Perez-Balaguer A, Ciprián D. Association of increased osteopontin and splice variant-c mRNA expression with HER2 and triple-negative/basal-like breast carcinomas subtypes and recurrence. Hum Pathol 2014;45:504-12. [Crossref] [PubMed]
- Lu S, Simin K, Khan A, et al. Analysis of Integrin β4 Expression in Human Breast Cancer: Association with Basal-like Tumors and Prognostic Significance. Clin Cancer Res 2008;14:1050-8. [Crossref] [PubMed]
- Choi Y, Lee HJ, Jang MH, et al. Epithelial-mesenchymal transition increases during the progression of in situ to invasive basal-like breast cancer. Hum Pathol 2013;44:2581-9. [Crossref] [PubMed]
- Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 2006;9:121-32. [Crossref] [PubMed]
- Wang Y, Cortez D, Yazdi P, et al. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev 2000;14:927-39. [PubMed]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet 2007;8:735-48. [Crossref] [PubMed]
- Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 2006;295:2492-502. [Crossref] [PubMed]
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol 2007;8:275-83. [Crossref] [PubMed]
- Xu X, Wagner KU, Larson D, et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet 1999;22:37-43. [Crossref] [PubMed]
- Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 2004;10:5367-74. [Crossref] [PubMed]
- Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005;5:341-54. [Crossref] [PubMed]
- Rimawi MF, Shetty PB, Weiss HL, et al. Epidermal growth factor receptor expression in breast cancer association with biologic phenotype and clinical outcomes. Cancer 2010;116:1234-42. [Crossref] [PubMed]
- Gauthier ML, Berman HK, Miller C, et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell 2007;12:479-91. [Crossref] [PubMed]
- Agarwal R, Gonzalez-Angulo AM, Myhre S, et al. Integrative analysis of cyclin protein levels identifies cyclin b1 as a classifier and predictor of outcomes in breast cancer. Clin Cancer Res 2009;15:3654-62. [Crossref] [PubMed]
- Keyomarsi K, Tucker SL, Buchholz TA, et al. Cyclin E and survival in patients with breast cancer. N Engl J Med 2002;347:1566-575. [Crossref] [PubMed]
- Voduc D, Nielsen TO, Cheang MC, et al. The combination of high cyclin E and Skp2 expression in breast cancer is associated with a poor prognosis and the basal phenotype. Hum Pathol 2008;39:1431-7. [Crossref] [PubMed]
- Saal LH, Holm K, Maurer M, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res 2005;65:2554-9. [Crossref] [PubMed]
- Saal LH, Gruvberger-Saal SK, Persson C, et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet 2008;40:102-7. [Crossref] [PubMed]
- Marty B, Maire V, Gravier E, et al. Frequent PTEN genomic alterations and activated phosphatidylinositol 3-kinase pathway in basal-like breast cancer cells. Breast Cancer Res 2008;10:R101. [Crossref] [PubMed]
- Shen WH, Balajee AS, Wang J, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 2007;128:157-70. [Crossref] [PubMed]
- Kamradt MC, Chen F, Cryns VL. The small heat shock protein αB-crystallin negatively regulates cytochrome c- and caspase-8-dependent activation of caspase-3 by inhibiting its autoproteolytic maturation. J Biol Chem 2001;276:16059-63. [Crossref] [PubMed]
- Moyano JV, Evans JR, Chen F, et al. AlphaB-crystallin is a novel oncoprotein that predicts poor clinical outcome in breast cancer. J Clin Invest 2006;116:261-70. [Crossref] [PubMed]
- Ivanov O, Chen F, Wiley EL, et al. αB-Crystallin is a novel predictor of resistance to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat 2008;111:411-7. [Crossref] [PubMed]
- Storci G, Sansone P, Trere D, et al. The basal-like breast carcinoma phenotype is regulated by SLUG gene expression. J Pathol 2008;214:25-37. [Crossref] [PubMed]
- DiMeo TA, Anderson K, Phadke P, et al. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res 2009;69:5364-73. [Crossref] [PubMed]
- Mani SA, Yang J, Brooks M, et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci USA 2007;104:10069-74. [Crossref] [PubMed]
- Su X, Malouf GG, Chen Y, et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget 2014;5:9864-76. [Crossref] [PubMed]
- Liu J, Shen L, Yao J, et al. Forkhead box C1 promoter upstream transcript, a novel long non-coding RNA, regulates proliferation and migration in basal-like breast cancer. Mol Med Rep 2015;11:3155-9. [Crossref] [PubMed]
- Zhuang Y, Nguyen HT, Burow ME, et al. Elevated expression of long intergenic non-coding RNA HOTAIR in a basal-like variant of MCF-7 breast cancer cells. Mol Carcinog 2015;54:1656-67. [Crossref] [PubMed]
- Khordadmehr M, Shahbazi R, Ezzati H, et al. Key microRNAs in the biology of breast cancer; emerging evidence in the last decade. J Cell Physiol 2019;234:8316-26. [Crossref] [PubMed]
- Bahmanpour Z, Sheervalilou R, Choupani J, et al. A new insight on serum microRNA expression as novel biomarkers in breast cancer patients. J Cell Physiol 2019;234:19199-211. [Crossref] [PubMed]
- Enerly E, Steinfeld I, Kleivi K, et al. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One 2011;6:e16915. [Crossref] [PubMed]
- de Rinaldis E, Gazinska P, Mera A, et al. Integrated genomic analysis of triple-negative breast cancers reveals novel microRNAs associated with clinical and molecular phenotypes and sheds light on the pathways they control. BMC Genomics 2013;14:643. [Crossref] [PubMed]
- Mouw JK, Yui Y, Damiano L, et al. Tissue mechanics modulate microRNA-dependent PTEN expression to regulate malignant progression. Nat Med 2014;20:360-7. [Crossref] [PubMed]
- Fonseca-Sanchéz MA, Pérez-Plasencia C, Fernández-Retana J, et al. microRNA-18b is upregulated in breast cancer and modulates genes involved in cell migration. Oncol Rep 2013;30:2399-410. [Crossref] [PubMed]
- Kim K, Chadalapaka G, Lee SO, et al. Identification of oncogenic microRNA-17-92/ZBTB4/specificity protein axis in breast cancer. Oncogene 2012;31:1034-44. [Crossref] [PubMed]
- Gong C, Qu S, Liu B, et al. MiR-106b expression determines the proliferation paradox of TGF-β in breast cancer cells. Oncogene 2015;34:84-93. [Crossref] [PubMed]
- Liu Y, Li H, Feng J, et al. Lin28 induces epithelial-to-mesenchymal transition and stemness via downregulation of let-7a in breast cancer cells. PLoS One 2013;8:e83083. [Crossref] [PubMed]
- Botti G, Cantile M. Circulating long non-coding RNAs: could they be a useful tool for cancer therapy monitoring? Expert Rev Anticancer Ther 2018;18:1167-8. [Crossref] [PubMed]


