
Sideroflexin1 as a novel tumor marker independently predicts survival in lung adenocarcinoma
Introduction
Lung cancer is the primary cause of cancerous death among men and the second leading cause of cancerous death among women around the world (1). There are tens of thousands of studies on ways to overcome lung cancer, and there are many prognostic factors associated with lung cancers survival. Targeted therapy based on these prognostic genes has made progress but needs more investigation.
The Sideroflexin (SFXN) family are mitochondrial tricarboxylate carrier proteins, that exist in the mitochondrial membrane (2). This family consists of five closely related members. A series of previous studies observed that SFXNs are associated with metabolism (3,4) and oral squamous cell carcinoma. SFXN2 participates in mitochondrial iron metabolism via the regulation of heme biosynthesis (3) and may contribute to blood pressure regulation and the plaque burden in the carotid artery (5,6). It was found that the serum levels of anti-SFXN3-autoAb increased markedly in patients with oral squamous cancer compared to normal controls. Also, the anti-SFXN3-autoAb serum level could be a potential biomarker during early screening for oral squamous cancer (7). SFXN3 also functions as a new mitochondrial protein regulating synaptic morphology in vivo (8). SFXN4 was identified as a candidate susceptibility gene for common familial colorectal cancer (9). Mutations in SFXN4 are a cause of mitochondriopathy and macrocytic anemia (10). SFXN5 does not correspond to PARK3 (2).
As for SFXN1, previous studies showed that it is significantly regulated in synovitis of osteoarthritis (OA) (11). In diffuse infiltrating gliomas, SFXN1 participates in an optimized gene subset associated with survival differences (12). Recent studies found that SFXN1 functions in the one-carbon metabolism process as a mitochondrial serine transporter (4). Indeed, one-carbon units sourced from serine contribute to cell proliferation in many carcinomas. Therefore, mitochondrial serine catabolism enzymes are commonly up-regulated in cancers. SFXN1 may play an important role in cancer but there have still not been many studies on it. In order to screen for novel prognostic biomarkers for lung adenocarcinoma (LUAD), we examined SFXN1.
In our research, we intend to show the discrepancy between SFXN1 mRNA and protein expression in LUAD tissues and paracancerous normal samples and demonstrate the prognostic value of SFXN1 mRNA expression in LUAD patients by analyzing data from a large patient cohort in TCGA and GEO. The possible reasons for SFXN1 dysregulation will also be investigated. Our research data will provide a basis for further functional exploration of this gene.
Methods
Data collection in TCGA and GEO
Level-3 data including gene expression and clinical information of LUAD patients in TCGA and GEO were acquired by the UCSC Xena Browser (https://xenabrowser.net/) and NCBI (https://www.ncbi.nlm.nih.gov/geoprofiles/). SFXN1 RNA-seq of 513 LUAD patients and 59 normal controls were downloaded from TCGA. The clinicopathological information of these patients, such as age, gender, smoking history, clinical stage, lymph nodal invasion status, residual tumors, survival status, overall survival (OS) in days, recurrence status, and recurrence-free survival (RFS) in days, were also downloaded. Five hundred cases had survival data records. The SFXN1 DNA copy number alteration (CNA) data (GISTIC2-processed) and DNA methylation data (measured by an Illumina 450k Infinium methylation BeadChip) were also downloaded through the UCSC Xena Browser. In GEO, the OS data of 115 patients with LUAD were downloaded.
SFXN1 protein immunofluorescence and immunohistochemistry staining
Immunohistochemistry (IHC) staining data of the SFXN1 protein came from the Human Protein Atlas (HPA; http://www.proteinatlas.org/). This is a large-scale protein research project whose main purpose is to map the locations of proteins encoded by genes expressed in human tissues and cells (13,14). Staining data, including those for lung tissues and LUAD and lung squamous cell carcinoma (LUSC) tissues were downloaded. An immunofluorescence analysis antibody with green fluorescence to SFXN1 was used to detect SFXN1 proteins, and immunofluorescence allowed the location detection of SFXN1 protein expression.
Statistical analysis
Data management and analysis were carried out with GraphPad Prism 7.0 and SPSS 24.0 software. The significance of SFXN1 expression differences among groups with different clinicopathological parameters was assessed through Welch’s unequal variances t-test. The median served as the cut-off for SFXN1 expression in Kaplan-Meier curves. The significance of differences between the survival curves was assessed with a log-rank test, and χ2 tests were utilized for assessing the statistical difference in clinicopathological parameters between the high and low SFXN1 expression groups. Univariate and multivariate Cox regression analyses were performed to evaluate whether unregulated SFXN1 expression independently predicts OS and RFS in LUAD patients. A P value <0.05 was considered statistically significant.
Results
SFXN1 mRNA expression was markedly upregulated in LUAD
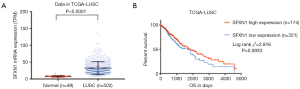
With the SFXN1 RNA-seq data consisting of 513 LUAD tissues and 59 paracancerous normal tissues from TCGA, we evaluated the mRNA expression discrepancies between the two groups. It was showed that SFXN1 was markedly more upregulated in LUAD tissues than in paracancerous normal tissues (Figure 1A,B). By detecting cellular immunofluorescence, we found that SFXN1 protein was localized in the cytoplasm (Figure 1C). According to SFXN1 IHC staining data in the HPA, normal lung tissues usually showed a weak degree of SFXN1 staining (Figure 1D). In contrast, among 11 cases of non-small cell lung cancer tissues examined, 1 case had negative SFXN1 staining (Figure 1E), 7 cases had moderate SFXN1 staining (Figure 1E), and the other 3 cases had strong SFXN1 staining (Figure 1E). We also evaluated the SFXN1 mRNA expression in LUSC patients from TCGA. The results indicated significantly upregulated SFXN1 expression in LUSC tissues compared to paracancerous normal tissues (P<0.0001; Figure S1A).
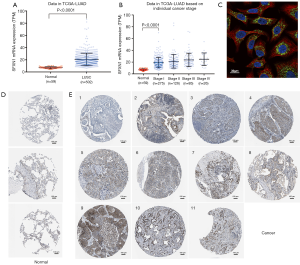
Upregulated SFXN1 mRNA expression predicted worse survival in LUAD patients
To confirm whether SFXN1 mRNA expression was associated with survival prognosis in LUAD patients, we extracted and analyzed survival information in both TCGA and GEO. In both databases, SFXN1 had markedly higher mRNA expression in the death group than the living group (Figure 2A,B). We also found that patients with positive lymph nodal invasion maintained increased SFXN1 mRNA expression (Figure 2C,D). Kaplan-Meier curves of OS were generated with data from TCGA and GEO, and the data indicated significantly worse OS in the high SFXN1 expression group compared to the low SFXN1 mRNA expression group (P=0.0015 and P=0.0207 respectively; Figure 2E,2F). Kaplan-Meier curves of RFS were also generated with data from TCGA, and we confirmed more favorable RFS in the low SFXN1 mRNA expression group than in the high SFXN1 mRNA expression group (Figure 2G). The statistical data on SFXN1 mRNA expression and the clinicopathological parameters from LUAD patients are arranged and summarized in Table 1. It was shown that patients in the high SFXN1 expression group had an obviously higher clinical stage (58 of 140 vs. 48 of 253; P=0.0004; Table 1). Furthermore, Kaplan-Meier curves of OS were generated for LUSC patients, and no significant correlation was found between the SFXN1 mRNA expression data and the OS data even under the best cut-off model (P=0.0933; Figure S1B).
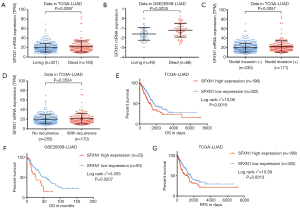
Table 1
| Parameters | SFXN1 expression | χ2 | P | |
|---|---|---|---|---|
| High (n=198) | Low (n=302) | |||
| Age, years (mean ± SD) | 64.42±0.7303 | 65.81±0.578 | – | 0.1328 |
| Gender | 3.313 | 0.0688 | ||
| Female | 97 | 173 | ||
| Male | 101 | 129 | ||
| Smoking history | 0.1829 | 0.6689 | ||
| 2/3/4/5 | 169 | 246 | ||
| 1 | 27 | 44 | ||
| No data | 2 | 12 | ||
| Clinical stage | 12.72 | 0.0004 | ||
| III/IV | 58 | 48 | ||
| I/II | 140 | 253 | ||
| Discrepancy/no data | 0 | 1 | ||
| Nodal invasion | 3.478 | 0.0622 | ||
| N0 | 119 | 205 | ||
| N1/2/3 | 75 | 90 | ||
| NX/no data | 4 | 7 | ||
| Residual tumors | 0.8945 | 0.3443 | ||
| R0 | 128 | 207 | ||
| R1/R2 | 8 | 8 | ||
| RX/no data | 62 | 87 | ||
| Radiation therapy | 2.2 | 0.1380 | ||
| No | 47 | 94 | ||
| Yes | 7 | 6 | ||
| No data | 144 | 202 | ||
| Recurrence status | 0.3957 | 0.5293 | ||
| No | 98 | 155 | ||
| Yes | 69 | 96 | ||
| No data | 31 | 51 | ||
| Living status | 3.56 | 0.0592 | ||
| Living | 116 | 202 | ||
| Dead | 82 | 100 | ||
| No data | 0 | 0 | ||
Smoking history: 1, lifelong non-smoker; 2, current smoker; 3, current reformed smoker (for >15 years); 4, current reformed smoker (for 15 years); 5, current reformed smoker (duration not specified). R0, no residual tumor; R1, microscopic residual tumor; R2, macroscopic residual tumor; RX, the presence of residual tumor cannot be assessed.
Upregulated SFXN1 mRNA expression independently predicts poor OS and RFS in LUAD patients
By performing univariate and multivariate analyses, we further evaluated whether upregulated SFXN1 mRNA expression independently predicts survival in LUAD patients. In the univariate analysis, it was indicated that advanced clinical stages, positive lymph nodal invasion, residual tumors and increased SFXN1 mRNA expression were correlated with unfavorable OS and RFS in patients with LUAD (Tables 2,3). With multivariate analysis we then confirmed that upregulated SFXN1 mRNA expression independently predicts unfavorable OS (HR: 1.621; 95% CI: 1.149–2.287; P=0.006) and RFS (HR: 1.678; 95% CI: 1.189–2.369; P=0.003) (Tables 2,3).
Table 2
| Parameters | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | ||
| Age (continuous) | 1.216 | 0.9068–1.629 | 0.4565 | – | – | – | |
| Gender (female vs. male) | 0.9534 | 0.7123–1.276 | 0.7471 | – | – | – | |
| Smoking history (2/3/4/5 vs. 1) | 0.9127 | 0.6065–1.376 | 0.6521 | – | – | – | |
| Clinical stage (III/IV vs. I/II) | 2.56 | 1.73–3.787 | <0.0001 | 1.672 | 1.096–2.552 | 0.017 | |
| Nodal invasion (positive vs. negative) | 2.547 | 1.846–3.515 | <0.0001 | 1.819 | 1.239–2.672 | 0.002 | |
| Residual tumors (yes vs. no) | 3.886 | 1.363–11.08 | <0.0001 | 2.522 | 1.349–4.714 | 0.004 | |
| SFXN1 expression (continuous) | 1.593 | 1.172–2.165 | 0.0015 | 1.621 | 1.149–2.287 | 0.006 | |
Smoking history: 1, lifelong non-smoker; 2, current smoker; 3, current reformed smoker (for >15 years); 4, current reformed smoker (for 15 years); 5, current reformed smoker (duration not specified). OS, overall survival; LUAD, lung adenocarcinoma.
Table 3
| Parameters | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | ||
| Age (continuous) | 1.201 | 0.8961–1.61 | 0.4499 | – | – | – | |
| Gender (female vs. male) | 0.9258 | 0.6915–1.24 | 0.6025 | – | – | – | |
| Smoking history (2/3/4/5 vs. 1) | 0.8852 | 0.5847–1.34 | 0.5466 | – | – | – | |
| Clinical stage (III/IV vs. I/II) | 2.508 | 1.699–3.701 | <0.0001 | 1.645 | 1.067–2.538 | 0.024 | |
| Nodal invasion (positive vs. negative) | 2.502 | 1.816–3.449 | <0.0001 | 1.689 | 1.148–2.486 | 0.008 | |
| Residual tumors (yes vs. no) | 4.365 | 1.442–13.21 | <0.0001 | 2.519 | 1.329–4.777 | 0.005 | |
| SFXN1 expression (continuous) | 1.607 | 1.182–2.185 | 0.0013 | 1.678 | 1.189–2.369 | 0.003 | |
Smoking history: 1, lifelong non-smoker; 2, current smoker; 3, current reformed smoker (for >15 years); 4, current reformed smoker (for 15 years); 5, current reformed smoker (duration not specified). RFS, recurrence-free survival; LUAD, lung adenocarcinoma.
DNA CNAs and DNA methylation could be the mechanisms of dysregulated SFXN1 mRNA expression in LUAD patients
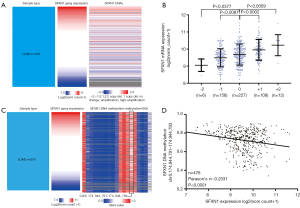
By examining SFXN1 DNA CNAs in 512 LUAD cases, we detected that copy deletion (−2 and −1; 32.03%) and copy amplification (+1 and +2; 23.63%) were frequent (Figure 3A,B), which might be important causes of dysregulated SFXN1 expression. Copy amplification (+1 and +2; 23.63%) was associated with significantly increased SFXN1 mRNA expression compared with the no change cases (0, P<0.01). In comparison, copy deletion (−2 and −1; 32.03%) was associated with significantly decreased SFXN1 mRNA expression (P<0.05). We next examined the methylation status of 19 CpG sites in SFXN1 DNA and discovered that the methylation status of one CpG site (black box) was negatively related to SFXN1 mRNA expression (Figure 3C,D), suggesting that DNA methylation may also contribute to dysregulated SFXN1 mRNA expression in LUAD patients. A higher level of DNA methylation was associated with decreased SFXN1 mRNA expression (P<0.01).
Discussion
Communication and coordination between mitochondria and the cell play an important role in various cell functions, such as metabolism, multiplication and survival (15). An imbalance in ell energetics is one of the important signals of cancer (16,17). Several potential mechanisms of deregulated cellular energetics are concerned with mitochondrial dysfunction caused by flawed mitochondrial enzymes, mutational mitochondrial DNA, or altered oncogenic signaling (18). Indeed, mitochondrial redox homeostasis control and biogenesis are often upregulated in cancer (19). While nuclear-encoded mitochondrial enzymes responsible for the tricarboxylic acid (TCA) cycle have mutations and generate oncogenic metabolites, human tumors are almost always malignant (20,21). Thus, mitochondria play a central, multidimensional role in the progression of therioma, and targeting mitochondria might be an effective therapeutic strategy (22).
Mitochondria play a central role in the metabolism of one-carbon units (23). One-carbon metabolism focuses on both the folate and methionine cycles and generates one-carbon units (methyl units) (24). One-carbon units are essential for nucleotide synthesis, reductive metabolism and methylation, and cancer cells require these pathways to support a high proliferation rate (25). Therefore, anti-folates, drugs targeting one-carbon metabolism, have been used for cancer treatment for a long time (26,27). The folate analog aminopterin acted as the first drug for leukemia relief (28), and methotrexate is still a mainstay in the management of acute lymphocytic leukemia (29). It was recently discovered that pemetrexed could function as a first-line drug for lung cancer (30). One-carbon metabolism consists of cytosolic and mitochondrial branches, and SFXN1 is a key component because it enables serine to enter mitochondria (4). Amino acids such as serine are a main one-carbon source, and cancerous cells are particularly susceptible to the loss of one-carbon units because of serine restriction or synthesis inhibition (25). Based on these findings, we can infer that SFXN1 may be closely associated with cancer progression. Some previous researches indicated that SFXN1 is abnormally expressed and could act as a pathogenic gene in diffuse infiltrating gliomas. Aberrations of SFXN1 are associated with unfavorable survival in patients with diffuse infiltrating gliomas (12).
In this study, we firstly found that SFXN1 had higher mRNA expression in LUAD than in normal controls. Then analyzing the long-term survival data of large LUAD cohorts from TCGA and GEO, we discovered significantly worse OS and RFS in patients with high SFXN1 mRNA expression. In addition, with univariate and multivariate analyses, we confirmed that upregulated SFXN1 mRNA expression independently predicted unfavorable OS (HR: 1.621; 95% CI: 1.149–2.287; P=0.006) and RFS (HR: 1.678; 95% CI: 1.189–2.369; P=0.003) among LUAD patients. In comparison, although SFXN1 was upregulated in LUSC tissues, no prognostic value associated with OS and RFS was found in LUSC patients. These findings indicate that SFXN1 could act as a valuable and specific prognostic biomarker in patients with LUAD.
SFXN1 may participate in an iron handling gene signature associated with survival in diffuse infiltrating gliomas (12). However, the specific mechanism of SFXN1 warrants a more pointed and specific investigation. Mechanistically, recent studies have shown that SFXN1 could be the main mitochondrial serine transporters in human cells and SFXN1 is expressed in some cancers, most highly in leukemias and lymphomas (4). Thus, SFXN1 becomes an important node in regulating the fate of serine and plays unexplored roles in cancer cell growth.
Many studies have revealed associations between DNA CNA and gene expression, as well as the changing methylation status of CpG islands and gene expression. In this research, we further investigated the possible reasons of SFXN1 dysregulation in patients with LUAD. The results showed that DNA CNAs and DNA methylation are possibly two significant reasons for SFXN1 overexpression in LUAD. Above all, genetic and epigenetic alterations frequently appear in LUAD patients and may have a profound effect on tumor phenotypes and patient survival. Consequently, it is meaningful and necessary in the future to find out how the genetic and epigenetic alterations of SFXN1 affect cancer cell behaviors in LUAD. Also, the relationship between CNA/methylation and clinical pathological features or evaluation of the CNA/ methylation and OS and RFS should be performed in the future.
Conclusions
SFXN1 expression increased more in both mRNA and protein levels in LUAD patients than in paracancerous normal tissues. Upregulated SFXN1 mRNA expression independently predicts unfavorable OS and RFS in patients with LUAD. DNA CNAs and methylation may be two important reasons for dysregulated SFXN1 mRNA expression in LUAD.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.06.34). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol 2016;893:1-19. [Crossref] [PubMed]
- Lockhart PJ, Holtom B, Lincoln S, et al. The human sideroflexin 5 (SFXN5) gene: sequence, expression analysis and exclusion as a candidate for PARK3. Gene 2002;285:229-37. [Crossref] [PubMed]
- Mon EE, Wei FY, Ahmad RNR, et al. Regulation of mitochondrial iron homeostasis by sideroflexin 2. J Physiol Sci 2019;69:359-73. [Crossref] [PubMed]
- Kory N, Wyant GA, Prakash G, et al. SFXN1 is a mitochondrial serine transporter required for one-carbon metabolism. Science 2018; [Crossref] [PubMed]
- Pott J, Burkhardt R, Beutner F, et al. Genome-wide meta-analysis identifies novel loci of plaque burden in carotid artery. Atherosclerosis 2017;259:32-40. [Crossref] [PubMed]
- Li C, Kim YK, Dorajoo R, et al. Genome-Wide Association Study Meta-Analysis of Long-Term Average Blood Pressure in East Asians. Circ Cardiovasc Genet 2017;10:e001527. [Crossref] [PubMed]
- Murase R, Abe Y, Takeuchi T, et al. Serum autoantibody to sideroflexin 3 as a novel tumor marker for oral squamous cell carcinoma. Proteomics Clin Appl 2008;2:517-27. [Crossref] [PubMed]
- Amorim IS, Graham LC, Carter RN, et al. Sideroflexin 3 is an alpha-synuclein-dependent mitochondrial protein that regulates synaptic morphology. J Cell Sci 2017;130:325-31. [Crossref] [PubMed]
- Gylfe AE, Katainen R, Kondelin J, et al. Eleven candidate susceptibility genes for common familial colorectal cancer. PLoS Genet 2013;9:e1003876. [Crossref] [PubMed]
- Hildick-Smith GJ, Cooney JD, Garone C, et al. Macrocytic anemia and mitochondriopathy resulting from a defect in sideroflexin 4. Am J Hum Genet 2013;93:906-14. [Crossref] [PubMed]
- Huang H, Zheng J, Shen N, et al. Identification of pathways and genes associated with synovitis in osteoarthritis using bioinformatics analyses. Sci Rep 2018;8:10050. [Crossref] [PubMed]
- Weston C, Klobusicky J, Weston J, et al. Aberrations in the Iron Regulatory Gene Signature Are Associated with Decreased Survival in Diffuse Infiltrating Gliomas. PLoS One 2016;11:e0166593. [Crossref] [PubMed]
- Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [Crossref] [PubMed]
- Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 2010;28:1248-50. [Crossref] [PubMed]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell 2012;148:1145-59. [Crossref] [PubMed]
- Loo JM, Scherl A, Nguyen A, et al. Extracellular metabolic energetics can promote cancer progression. Cell 2015;160:393-406. [Crossref] [PubMed]
- Hsu CC, Tseng LM, Lee HC. Role of mitochondrial dysfunction in cancer progression. Exp Biol Med (Maywood) 2016;241:1281-95. [Crossref] [PubMed]
- Srinivasan S, Guha M, Kashina A, et al. Mitochondrial dysfunction and mitochondrial dynamics-The cancer connection. Biochim Biophys Acta Bioenerg 2017;1858:602-14. [Crossref] [PubMed]
- Marengo B, Nitti M, Furfaro AL, et al. Redox Homeostasis and Cellular Antioxidant Systems: Crucial Players in Cancer Growth and Therapy. Oxid Med Cell Longev 2016;2016:6235641. [Crossref] [PubMed]
- Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene 2006;25:4663-74. [Crossref] [PubMed]
- Singh AK, Pandey P, Tewari M, et al. Human mitochondrial genome flaws and risk of cancer. Mitochondrial DNA 2014;25:329-34. [Crossref] [PubMed]
- Zong WX, Rabinowitz JD, White E. Mitochondria and Cancer. Mol Cell 2016;61:667-76. [Crossref] [PubMed]
- Lan X, Field MS, Stover PJ. Cell cycle regulation of folate-mediated one-carbon metabolism. Wiley Interdiscip Rev Syst Biol Med 2018;10:e1426. [Crossref] [PubMed]
- Ducker GS, Rabinowitz JD. One-Carbon Metabolism in Health and Disease. Cell Metab 2017;25:27-42. [Crossref] [PubMed]
- Newman AC, Maddocks ODK. One-carbon metabolism in cancer. Br J Cancer 2017;116:1499-504. [Crossref] [PubMed]
- Konno M, Asai A, Kawamoto K, et al. The one-carbon metabolism pathway highlights therapeutic targets for gastrointestinal cancer Int J Oncol 2017;50:1057-63. (Review). [Crossref] [PubMed]
- Cheung A, Opzoomer J, Ilieva KM, et al. Anti-Folate Receptor Alpha-Directed Antibody Therapies Restrict the Growth of Triple-negative Breast Cancer. Clin Cancer Res 2018;24:5098-111. [PubMed]
- Farber S, Diamond LK. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N Engl J Med 1948;238:787-93. [Crossref] [PubMed]
- Rudin S, Marable M, Huang RS. The Promise of Pharmacogenomics in Reducing Toxicity During Acute Lymphoblastic Leukemia Maintenance Treatment. Genomics Proteomics Bioinformatics 2017;15:82-93. [Crossref] [PubMed]
- Scagliotti GV, Kortsik C, Dark GG, et al. Pemetrexed combined with oxaliplatin or carboplatin as first-line treatment in advanced non-small cell lung cancer: a multicenter, randomized, phase II trial. Clin Cancer Res 2005;11:690-6. [PubMed]


