
Overexpression of CASC19 indicates poor prognosis and facilitates proliferation, migration and invasion in colorectal cancer
Introduction
CRC is the fifth most frequent cause of cancer deaths in China, with the incidence increasing continuously (1). Approximately 50% of patients with colorectal cancer develop colorectal liver metastases (CRLM), which are one of major causes of deaths (2). Though there were a lot of alterations in oncogenes and tumor suppressor genes in CRLM (3,4), the exact molecular mechanisms involved in CRLM pathogenesis remain unknown. The long noncoding RNAs (lncRNAs) are a kind of RNA molecules over 200 nucleotides, which could be transcribed but have no protein-coding capacity (5). Previous studies found that lncRNAs have a crucial role in carcinogenesis and distant metastasis through regulating gene expression (6-8). Aberrant lncRNA expression has been reported in different kinds of cancers, including colorectal cancers (9-12), suggesting that lncRNAs may be associated with tumorigenesis and tumor progression.
In colorectal cancer, a number of lncRNAs were associated with tumor progression or metastasis, such as HOTAIR (13). Nevertheless, for the functional roles of lncRNAs in CRLM, there is still few integrative analysis. LncRNA expression has higher tissue-specificity than protein-coding gene expression (14), which suggests that lncRNAs are potential biomarkers for CRLM. Therefore, identifying lncRNAs associated with CRLM might provide new insights into the pathogenesis of CRLM as well as potential biomarker candidates.
In this work, comprehensive analysis of the long noncoding transcriptome of CRLM and patient-matched specimens were performed to identify differentially expressed lncRNAs in normal tissues, tumor tissues and matched liver metastasis tissues. Furthermore, we used Short Time-series Expression Miner (STEM) and assessed the tissue specificity of the lncRNA expression and its relationship with CRLM. Finally, we examined in vitro effects of one long non-coding RNAs, termed CASC19, on tumorigenesis and tumor progression.
Methods
Patients and samples
This study included analysis of a total of 291 fresh tissue specimens from three cohorts of CRC. Cohort 1 (discovery cohort) included 4 patients with CRLM, which encompassed 12 samples of primary colorectal adenocarcinoma tumor tissues (T), normal tissues (N) and matched liver metastasis tissues (M). Cohort 2 (validation cohort) enrolled 21 patients with CRLM, which was used to confirm the discovery by cohort 1. And a total of 108 CRC patients were enrolled into cohort 3 to determine relationship between prognosis and lncRNAs identified in cohort 1 and cohort 2. Written informed consent from each patient with CRC was obtained at the FUSCC.
Cell culture
We purchased the human embryonic kidney cell line 293T and the human CRC cell lines (HCT-116, LoVo, SW480, SW620, RKO, HT-29) from the Type Culture Collection of the Chinese Academy of Sciences. The HCT-116 and HT-29 cells, LoVo cells and 293T cells were respectively cultured in McCoy’s 5A, F12K medium, and Dulbecco’s modified Eagle medium, and were maintained at 37 °C containing 5% CO2. The other CRC cell lines were cultured in RPMI 1,640 medium. All media were supplemented with 10% fetal bovine serum and 100 U/mL penicillin/streptomycin (Gibco). Permanent stocks of purchased cells were prepared, and all cells were stored in liquid nitrogen. Cells were used for experiments within 6 months.
Total RNA extraction and cDNA synthesis
Total RNA was extracted from fresh specimens or cells by TRIzol reagent (Invitrogen, Life Technologies), and RNA was quantified by a NanoDrop spectrophotometer. Then RNA (500 ng) was reverse-transcribed by the PrimeScript™ RT Master Mix Kit (Takara Biotechnology). We amplified cDNA through PCR by SYBR Premix DimerEraser (Perfect Real Time; Takara Biotechnology), and real-time quantification PCR was performed using the Applied Biosystems 7900HT Fast Real-Time PCR System (Life Technologies).
Real-time quantitative reverse transcriptase polymerase chain reaction
qRT-PCR was performed using the 7900 Real-Time PCR System with the SDS 2.3 software sequence detection system (Applied Biosystems). The relative abundance of target transcripts was evaluated utilizing 5 ng of cDNA and SYBR Green (Takara), and the results were normalized to the expression levels of ACTB using the 2−ΔCt method; ΔCt is the difference of Ct values between the lncRNA of interest and the normalizer. For each sample, qRT-PCR was performed in duplicate and the mean value was used to calculate the expression levels. Normalized values were further log transformed and standardized.
Vector construction
The lncRNA CASC19 was amplified by polymerase chain reaction (PCR), and then the PCR products were digested and ligated into the pLVTHM vector to construct a CASC19-overexpression vector. The sequences of the primers and siRNA are listed in Table S1.
Migration and invasion assays
The invasion assay was carried out using a 24-well Millicell chamber containing a Matrigel-coated membrane. The migration assay was carried out in a similar fashion but without the Matrigel coating. Cells (5×105 cells in serum-free medium) were seeded in the top chamber. The bottom wells were filled with complete medium. Cells on the upper membrane surface were wiped off using a cotton swab, and the lower membrane surface was fixed with methanol, stained with 0.1% crystal violet, and cells were counted in five random fields (original magnification, ×200).
Cell proliferation assay
We assessed cell proliferation by Cell Counting Kit (CCK)-8 (Donjindo) to estimate cell viability. Treated cells (i.e., overexpression and knocked down cell lines) were seeded into 96-well plates at an initial density of 1×103 cells/well. After 24, 48, 72, and 96 h of cultivation, CCK-8 solution was added to each well and incubated for 2 h. The absorbance was measured by scanning with a microplate reader (MRX; Dynex Technologies, West Sussex, UK) at 450 nm.
STEM analysis
The specific tissue expression profiles were determined by STEM analysis. The values of lncRNA expression of tumor tissue were represented as log ratios relative to the expression of normal tissue. Then the approach selects a set of predetermined temporal model profiles and determines the statistical significance of the number of genes assigned to each profile compared to the number of genes expected based on chance.
Statistical analysis
We performed all statistical analyses using SPSS software. Error bars in figures represent the standard deviation. Expression levels of each lncRNA were compared by the paired Mann-Whitney test. The clinicopathological characteristics were compared between the two groups using the Fisher exact test or χ2 test. Statistical significance was determined using the Student t-test. OS curves were analyzed using the Kaplan-Meier method, with P value determined by a log-rank test. Hazard ratios (HRs) and 95.0% confidence intervals (CIs) were calculated. A P value of <0.05 was regarded as statistically significant.
Results
The long noncoding transcriptome profile of CRLM
To investigate the association between CRLM and lncRNAs expression, we applied RNA-seq technique to analyze cohort 1, which encompassed 12 samples of primary colorectal adenocarcinoma tumor tissues (T), normal tissue (N) and matched liver metastasis tissue (M) from 4 patients with CRLM.
We organized the lncRNAs according to their expression profiles in 3 different types of tissues. Figure 1A showed the overall lncRNA expression profiles. Distinct groups of lncRNAs were expressed in 3 different types of tissues. The sequencing data of 3 tissues were normalized and the specific tissue gene expression profiles were identified by STEM analysis. Seven lncRNA profiles of total 16 model profiles showed significant difference (Figure 1B). The change of lncRNA expression in T, N and M tissues was represented by the black lines in the profile boxes. The profile number on the top corner of each profile box presented each model profile and the number on the bottom represented the adjusted p-value of lncRNAs expression across 3 different types of tissues. We focus on continuous up-regulation patterns in 3 types of tissues such as profiles 12, 13 and 15. Finally, we identified 63 lncRNA according with the trend of N < T < M.
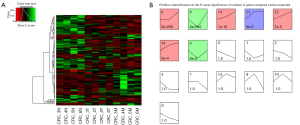
Identification of novel CRLM related lncRNAs
We evaluated the 63 lncRNAs expression by qRT-PCR analysis in cohort 2, which included 21 patients with CRLM. The results validated the expression trend of N < T < M in 9 candidate lncRNAs (Figure 2). Five candidate lncRNAs were excluded for subsequent analysis, because of low expression level (e.g., HL-12) or no significantly difference among N, T and M tissues (e.g., HL-3, HL-10, HL-21, HL-23).Finally, four lncRNAs (HL-5, HL-20, HL-38, and HL-59) with the trend of N < T < M were analyzed in the subsequent screening experiments.
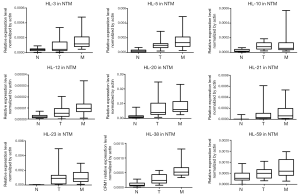
Up-regulation of CASC19 is associated with CRLM and poor prognosis
Then we detected the expression of these 4 lncRNAs in 108 colorectal cancer tissues using qRT-PCR, and analyzed the prognosis value of these lncRNAs. The three lncRNAs (HL-5, HL-20 and HL-38) had no associated with overall survival. However, high expression of HL-59 (CASC19) was associated with poor prognosis (P=0.013, Figure 3). Furthermore, the patients were divided into high (n=32) and low (n=76) CASC19 expression groups according to the receiver operating characteristic curve of CASC19 levels. Univariate analyses showed patients with high CASC19 expression had a poorer prognosis compared to patients with low CASC19 expression (P=0.013, Table 1). The CASC19 aberrant high expression level was significantly correlated with distant metastasis (P=0.026) and mid-highly differentiated histological grade (P=0.018) (Table 2). However, there was no relationship between CASC19 expression and other factors such as sex (male vs. female), age (≤60 vs. >60), and lymph node metastasis (N0 vs. N1 or above).
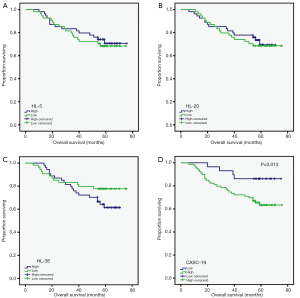
Table 1
| Variables | Case no. | Average OS (months) | P value |
|---|---|---|---|
| Age | 1 | ||
| >60 | 51 | 60.9 | |
| ≤60 | 57 | 60.9 | |
| Sex | 0.594 | ||
| Male | 66 | 61.4 | |
| Female | 42 | 61.2 | |
| Tumor type | 0.062 | ||
| Colon cancer | 63 | 57.3 | |
| Rectal cancer | 45 | 67.2 | |
| Tumor size | 0.306 | ||
| ≥5 | 43 | 55.4 | |
| <5 | 65 | 63.7 | |
| Depth of invasion | 0.016 | ||
| T1/T2/T3 | 27 | 71.4 | |
| T4 | 81 | 58.2 | |
| Lymph node metastasis | 0.232 | ||
| Negative | 42 | 62.2 | |
| Positive | 66 | 58.8 | |
| Distant metastasis | <0.001 | ||
| Negative | 12 | 28.5 | |
| Positive | 96 | 66.1 | |
| Venous invasion | 0.017 | ||
| Negative | 39 | 62.8 | |
| Positive | 66 | 54.5 | |
| Perineural invasion | 0.065 | ||
| Negative | 84 | 63.7 | |
| Positive | 20 | 54.4 | |
| Histology type | 0.247 | ||
| Adenocarcinoma | 82 | 60.4 | |
| Others | 25 | 64.8 | |
| Differentiated degree | 0.104 | ||
| Poorly differentiated | 27 | 55.8 | |
| High-middle differentiated | 73 | 64.2 | |
| CASC19 | 0.013 | ||
| High expression | 76 | 55 | |
| Low expression | 32 | 70.5 |
Table 2
| Variables | CASC19 expression in CRC | P value | |
|---|---|---|---|
| High expression | Low expression | ||
| Differentiated degree | 0.018 | ||
| Poorly differentiated | 15 (20.8) | 12 (42.9) | |
| High-middle differentiated | 57 (79.1) | 16 (57.1) | |
| Distant metastasis | 0.026 | ||
| Positive | 12 (15.6) | 0 (0.0) | |
| Negative | 65 (84.4) | 31 (100.0) | |
Knockdown and overexpression of CASC19 influence proliferation, migration and invasion in vitro
HCT-116 and LoVo cell lines were used to analyze the function of CASC19. Figure 4A show that knockdown (with siRNA) and overexpression (with transfection of pLVTHM-CASC19) of CASC19 in these cell lines were successfully constructed (P<0.001). The growth curves determined by CCK-8 assays indicated that knockdown of endogenous CASC19 expression dramatically reduced cell proliferation in both HCT-116 and LoVo cells, whereas CASC19 overexpression enhanced the proliferative capacity in both cell lines (P<0.001, Figure 4B). Knockdown of CASC19 expression significantly inhibited the colony formation of HCT-116 and LoVo cells, whereas overexpression of CASC19 had the opposite effect (P<0.001, Figure 4C), which were shown in colony formation assays. Therefore these results collectively suggested that overexpression of CASC19 could enhance CRC cell growth.
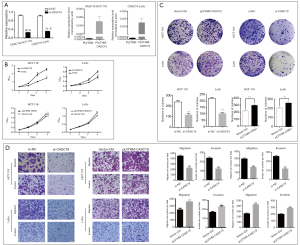
CASC19 knockdown inhibited cell migration more than in LoVo cells and HCT-116 cells (P<0.001), and significantly decreased cell invasion (P<0.001), as determined with the Matrigel assays. In contrast, cell invasion and migration were increased when CASC19 was overexpressed in HCT-116 and LoVo cells (P<0.001, Figure 4D). These results indicated that overexpression of CASC19 could facilitate CRC cell migration and invasion in vitro.
Discussion
A mass of lncRNAs encoded by human genes are dynamically expressed at different developmental stages, show highly specific expression patterns across different tissues, and play key roles in diverse biological processes (15). At present, a large number of lncRNAs have been identified and demonstrated to be involved in various types of cancers (16,17). In recent years, the usage of high-throughput technologies provides an easier way to profile the long noncoding transcriptome.
As far as we know our research is the first study to investigate the differences in the long noncoding transcriptome profile of primary colorectal cancer tissues, adjacent normal tissues and matched colorectal cancer liver metastasis using RNA-seq technique. The study identified several CRLM-associated lncRNAs, such as CCAT1, GAS5 and PVT1, which have been reported to be involved in tumor progression and biological pathways and some lncRNAs have not been previously reported. Furthermore, we used Short Time-series Expression Miner (STEM) and focused on the lncRNAs which exert the expression trend of N < T < M and might act as promoting effect on CRLM. We identified 63 lncRNAs according with the trend of N < T < M, and after validation in cohort 2, 5 lncRNAs were showed to be associated with CRLM. Then we evaluated the prognosis of these five lncRNAs in 108 CRC cohort, and high expression of CASC19 was correlated with poor overall survival of CRC.
In the present study, our results revealed that the expression of CASC19 increased in CRC tissues and matched liver metastasis tissues. CRC with higher CASC19 expression tended to be associated with high-middle differentiation state and distant metastasis. What’s more, survival analysis showed that CASC19 overexpression was significantly associated with poorer overall survival of CRC patients, which suggested CASC19 could serve as a potential biomarker for advanced CRC patients. The CCK-8 assays and Matrigel assays showed that overexpression of CASC19 could enhance CRC cell growth, migration and invasion. Overall, our results indicated that CASC19 may act as tumor promoter in CRC.
In conclusion, liver metastasis tissues and primary tissues of CRC showed different expression profiles of lncRNAs, and we identified five lncRNAs associated with CRLM. We suggest that CASC19 could serve as a promising biomarker associated with poor prognosis and liver metastasis in advanced CRC patients.
Table S1
| Primers and siRNA | Sequence (5'→3') |
|---|---|
| HL-3-F | AGACCCAGCGGCCAGCACA |
| HL-3-R | TCGGCACCCCTCCTTCCCTC |
| HL-5-F | TCTGGCGGTGTGCGCTGAG |
| HL-5-R | CCACCTTGTCCAGGTTTCAGGGA |
| HL-10-F | CGATCCACTCCCGGCTGTTCC |
| HL-10-R | CGGTGTTCAAGGGCAATTCCC |
| HL-12-F | GCGTTCTGAGGAACTGGTG |
| HL-12-R | AAATAATAGGCATGTGGTG |
| HL-20-F | TCTGATGGCAGCAACCACCTGG |
| HL-20-R | TGAACGGGGCCTTGGCGTC |
| HL-21-F | GAGGGGCTCTGGAGCAGAAAGAG |
| HL-21-R | CCGCTGTCAGGAAACTGTGTGGA |
| HL-23-F | CGAGTTGGGGGAGGCAGGTG |
| HL-23-R | CCAGCAGCAGCTCTTGTCTTGGG |
| HL-38-F | CCCAGCAATGGAGCCTAAGAAGG |
| HL-38-R | TGGCTACGTAATCTTCCCAGGGC |
| HL-59 (CASC19)-F | TGCCTGGGTTAGAACCCTGCTG |
| HL-59 (CASC19)-R | TGGAGGGTGCCATCACAGTCAGT |
| β-actin-F | TTGGCCAGGGGTGCTAAG |
| β-actin-R | AGCCAAAAGGGTCATCATCTC |
| CASC19 siRNA-1 | GCTCAGCATTTGCCATACT |
| CASC19 siRNA-2 | CCTTAGAATTGGAGTGCCT |
| pLVTHM-CASC19-F (BamH1) | CGGGATCC CTAGCTCAGCATTTGCCATACTACA |
| pLVTHM-CASC19-R (Ecor1) | CGGAATTC TTTGGACAGCACCTTGAATGCTATC |
Acknowledgments
Funding: This research was supported in part by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.07.17). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board Committee of the FUSCC (Shanghai, China) and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Moulton CA, Gu CS, Law CH, et al. Effect of PET before liver resection on surgical management for colorectal adenocarcinoma metastases: a randomized clinical trial. JAMA 2014;311:1863-9. [Crossref] [PubMed]
- Balachandran VP, Arora A, Gönen M, et al. A Validated Prognostic Multigene Expression Assay for Overall Survival in Resected Colorectal Cancer Liver Metastases. Clin Cancer Res 2016;22:2575-82. [Crossref] [PubMed]
- Ki DH, Jeung HC, Park CH, et al. Whole genome analysis for liver metastasis gene signatures in colorectal cancer. Int J Cancer 2007;121:2005-12. [Crossref] [PubMed]
- Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res 2011;71:3-7. [Crossref] [PubMed]
- Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer?. Hum Mol Genet 2010;19:R152-61. [Crossref] [PubMed]
- Gómez-Maldonado L, Tiana M, Roche O, et al. EFNA3 long noncoding RNAs induced by hypoxia promote metastatic dissemination. Oncogene 2015;34:2609-20. [Crossref] [PubMed]
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6. [Crossref] [PubMed]
- Gutschner T, Hämmerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 2013;73:1180-9. [Crossref] [PubMed]
- Fan Y, Shen B, Tan M, et al. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res 2014;20:1531-41. [Crossref] [PubMed]
- Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 2010;38:5366-83. [Crossref] [PubMed]
- Ji Q, Zhang L, Liu X, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer 2014;111:736-48. [Crossref] [PubMed]
- Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011;71:6320-6. [Crossref] [PubMed]
- Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011;25:1915-27. [Crossref] [PubMed]
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013;154:26-46. [Crossref] [PubMed]
- Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol 2011;29:742-9. [Crossref] [PubMed]
- Brunner AL, Beck AH, Edris B, et al. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol 2012;13:R75. [Crossref] [PubMed]


