The influence of tumor heterogeneity on sensitivity of EGFR-mutant lung adenocarcinoma cells to EGFR-TKIs
Introduction
Lung cancer is the leading cause of cancer-related death worldwide, with non-small cell lung cancer (NSCLC) being the most common type (1). The 5-year survival rate of patients with NSCLC is only approximately 15% (2). Previous studies have identified some activating mutations (i.e., sensitive mutations) in the epidermal growth factor receptor (EGFR) as biomarkers which predicted a good response to tyrosine kinase inhibitors of EGFR (EGFR-TKIs), such as erlotinib and gefitinib (3). Many randomized controlled trials have shown that patients with EGFR-sensitive mutations [especially a deletion in exon 19 (E19del) and a point mutation in exon 21 L858R (E21 L858R)] who received EGFR-TKI treatment showed a significantly higher overall response rate of about 70% and longer median progression-free survival of 9–13 months compared to those who received chemotherapy alone (4-7). However, most patients who respond to EGFR-TKIs will eventually develop secondary resistance after a period of response; in addition, approximately 20% of patients show initially poor sensitivity (i.e., primary resistance) to EGFR-TKIs.
There are two critical mechanisms involved in the secondary resistance of EGFR-TKIs: one is acquisition of a mutation of the EGFR, most commonly seen in EGFR exon 20 T790M, another is the activation of a bypass signaling pathway of EGFR, such as amplification of the MET or HER2 gene, or epithelial–mesenchymal transition (8,9). Recent studies have shown that extensive genetic heterogeneity or so-called spatial heterogeneity is present within tumors (10). Due to the genetic instability and evolution during clonal proliferation of tumor cells, the EGFR-sensitive mutant type of tumor cells may co-exist with other subclonal tumor cells such as EGFR-TKI-resistant mutants or wild-type tumor cells within tumors when the tumor occurs. Tumor heterogeneity may be associated with primary resistance and secondary resistance to EGFR-TKIs. Using a highly sensitive detection method, researchers found that so-called resistant subclonal variations such as EGFR T790M or MET amplification could co-exist with EGFR-sensitive mutant type at a low-copy number in tumors before EGFR-TKI treatment. After EGFR-TKI treatment, the sensitive mutant lung cancer cells are suppressed, so the EGFR-TKI-resistant mutant lung cancer cells become the main clone. This suggests that “clonal selection” for EGFR-TKI-resistant mutations may lead to secondary resistance to EGFR-TKIs (11,12). Zhou et al. found that patients with a high abundance of EGFR mutations may benefit more than those with a low abundance (13). However, the exact mechanism underlying the involvement of heterogeneity in EGFR-TKI resistance is not yet clear.
Besides the above-mentioned intratumoral heterogeneity, the tumor microenvironment (TME), which contains microvessels, immune cells, fibroblasts and other stromal components surrounding the tumor cells, may interact with tumor cells by paracrine action to regulate cell proliferation, invasion, angiogenesis and metastasis (9). Large amounts of hepatocyte growth factor (HGF) are secreted by fibroblasts in the TME and can activate the downstream PI3K-AKT signal pathway by combining with its receptor MET in the tumor cell membrane (11). Thus, crosstalk between stromal cells in the TME and intratumoral EGFR-sensitive mutant lung cancer cells may be involved in the primary or secondary resistance of EGFR-TKIs. If so, even though the EGFR pathway is blocked, activation of a bypass signal pathway may also promote tumor cell proliferation.
In the present study, we simulated the tumor heterogeneity using transwell co-culture technology to explore the influence of heterogeneous tumor cells or stromal cells in the TME on the sensitivity of EGFR-sensitive mutant lung cancer cells to EGFR-TKIs and to investigate relevant molecular mechanisms.
Methods
Cells and reagents
Human lung adenocarcinoma cell lines including the cell line H1975 with EGFR E20 T790M, the cell line PC-9 with EGFR E19 E746-A750 del, and the A549 cell line with wild type EGFR, as well as the human lung embryonic fibroblast cell line MRC-5, were obtained from West China Hospital of Sichuan University (Chengdu, China). Erlotinib was purchased from Santa Cruz Biotechnology, Inc. (CA, USA). ELISA detection kits for HGF, transforming growth factor-α (TGF-α), and insulin-like growth factor-1 (IGF-1) were purchased from USCN Life Science Inc. (Shanghai, China). High-purity total RNA extraction kits were purchased from Generay Biotech Co., Ltd. (Shanghai, China), RevertAid First Strand cDNA synthesis kits were purchased from Fermentas Inc. (Thermo Fisher Scientific, Waltham, MA, USA), and PCR reagent Super Real Premix Plus (with SYBR Green I) was purchased from Tiangen Biotech Co., Ltd. (Beijing, China). Primary antibodies against Met (c-Met) (EP1454Y), Met (c-Met) (phospho Y1349) (EP2367Y), ErbB3 (E186), EGFR (EP38Y) and EGFR (phospho Y1092) (EP774Y) were purchased from Abcam (Cambridge, UK). Akt (C67E7), phospho-Akt (Ser473) (D9E), ERK1/2 and phospho-ERK1/2 (Thr202/Tyr204) were purchased from Cell Signaling Technology (Danvers, MA, USA); PI3-kinase p110 & alpha (H-201) were purchased from Santa Cruz Biotechnology.
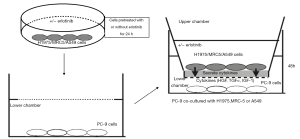
Cell co-culture
Three co-culture systems–H1975/PC-9, A549/PC-9, and MRC5/PC-9 (Thermo Fisher Scientific)–were established using the transwell technique, and grouped according to the different proportions of H1975/PC-9, A549/PC-9, and MRC5/PC-9 (as shown in Table 1 and Figure 1). Each co-culture system was then subgrouped to be treated with erlotinib (erlotinib+) or without erlotinib (erlotinib-). Specifically, cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM), supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/mL) and streptomycin (50 U/mL) in a humidified CO2 incubator at 37 °C. In preliminary experiments, we determined that the IC50 concentration of PC-9 cells to erlotinib at 72 h was 3 µM/L. Cells of A549, H1975 and MRC-5 during the exponential growth phase were rinsed in PBS, then pretreated in complete medium with or without erlotinib (3 µM/L) for 24 h. PC-9 cells (2×105/mL) treated with or without erlotinib (3 µM/L) were placed in DMEM complete medium, and 1 mL cell suspension was added to the bottom chamber. Then A549, H1975 and MRC-5 cells were suspended in DMEM complete medium with or without erlotinib (3 µM/L) at cell densities of 2×103/mL, 2×104/mL, 1×105/mL, 2×105/mL and 6×105/mL, and 1 mL of the cell suspension was added to the upper chamber. The co-cultures were then incubated at 37 °C, in a 5% CO2 saturated humidity incubator for 48 h. PC-9 cells cultured alone were used as the control group and the cell density, erlotinib concentration, and treatment time were all in accordance with the co-culture groups.
Table 1
| Chamber | Cells | Group 1 | Group 2 | Group 3 | Group 4 | Group 5 |
|---|---|---|---|---|---|---|
| Upper chamber | MRC5/H1975/A549 | 1% | 10% | 50% | 100% | 300% |
| Lower chamber | PC-9 | 1 | 1 | 1 | 1 | 1 |
| Intervention factor | Erlotinib | +/− | +/− | +/− | +/− | +/− |
PC-9 cell viability assay
For the PC-9 cell viability assay, the transwell upper chamber was removed, growth medium was aspirated from the lower chamber, and Cell Counting Kit-8 (CCK8) reagents diluted 1:100 in preheated DMEM complete medium was added at 1 mL per well. The plates were then incubated at 37 °C, in a 5% CO2 saturation humidity incubator, for 1h. After incubation, 100 µL of culture supernatant were transferred into wells of 96-well plates, and the absorbance (OD value) was measured with a microplate reader at a wavelength of 450 nm. Each experiment was repeated three times.
Determination of HGF, TGF-α and IGF-1 in co-culture supernatant
The levels of the cytokines HGF, IGF-1 and TGF-α in the PC-9 cell supernatants were determined after co-culture with various numbers of MRC-5, H1975, or A549 cells. The transwell systems were incubated for 48 h, and then the supernatant of the PC-9 cells was harvested and centrifuged. Each ELISA was performed according to the manufacturer’s recommended procedures (Immunis HGF EIA kit, Institute of Immunology, Japan). Color intensity was measured at 450 nm with a spectrophotometric plate reader. Growth factor concentrations were determined by comparison with standard curves. All samples were run in triplicate.
Detection of MET amplification by quantitative PCR
MET gene amplification of PC-9 cells was examined by real-time quantitative PCR (qRT-PCR). qRT-PCR was performed on genomic DNA purified with a RevertAid First Strand cDNA synthesis Kit. Primer sequences were as follows: Homo MET forward primer: 5'-TTGTAAGTGCCCGAAGTGTA-3'; reverse primer: 5'-TTTCTTGCCATCATTGTCC-3'. Homo GAPDH forward primer: 5'-AGAAGGCTGGGGCTCATTTG-3'; reverse primer: 5'-AGGGGCCATCCACAGTCTTC-3' (14). PCR reactions were carried out in triplicate with 20 ng genomic DNA, 900 nmol/L primers, and 250 nmol/L probes under standard thermocycling conditions (50.0 °C for 3 min, 95.0 °C for 15 min, then 45 cycles of 95.0 °C for 10 s, 61.5 °C for 20 s, 72.0 °C for 20 s followed by melting curve analysis from 70 to 95 °C at 0.5 °C increments).
Western blotting
Exponential phase cells were lysed for 30 min in cell lysis buffer (150 µL) containing PMSF (1 mM), then the lysate was centrifuged at 11,000 rpm, 4 °C for 5 min. The supernatant was stored at −80 °C until analysis. The protein concentrations were determined by the BCA method (Bicinchoninic acid), and 30 µg of protein was separated by 10% polyacrylamide gel electrophoresis, then transferred onto a PVDF membrane. The membrane was blocked with 5% skimmed milk powder or bovine serum albumin overnight, and then incubated for 2 h at 4 °C with one of the following primary antibodies: c-Met, phospho-c-Met, ErbB3, EGFR, phospho-EGFR, Akt, phospho-Akt, ERK1/2, phospho-ERK1/2, PI3-kinase. After washing three times, the membranes were incubated for 1 h at room temperature with the appropriate secondary antibody. Immunoreactive bands were visualized by imaging with enhanced chemiluminescence reagent (Beyotime Biotechnology Co., Shanghai, China). Pictures were exported to Image J software and used for further analysis of the optical density of every band.
Statistical analysis
The statistical significance of differences was analyzed by one-way ANOVA. All statistical analyses were performed using GraphPad Prism Ver. 6.0 software (GraphPad Software Company, La Jolla, CA, USA). P<0.05 was considered statistically significant.
Results
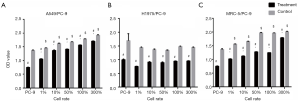
Cell viability of co-cultured PC-9 cells
In the three co-culture systems, the cell activity of PC-9 cells in the erlotinib+ subgroup decreased markedly compared with that in the erlotinib− subgroup, Pall<0.05. In co-culture systems of A549/PC-9 or MRC-5/PC-9, the cell viability of the PC-9 cells in the lower chamber gradually increased to various extents with the increase in the proportion of upper chamber cells relative to lower chamber cells from 1% to 300% in both erlotinib+ and erlotinib−subgroups, with statistical significance of Pall<0.05 compared with the PC-9 alone group. However, no significant change in cell viability was observed at any proportion of H1975 cells in the H1975/PC-9 co-culture system (Figure 2).
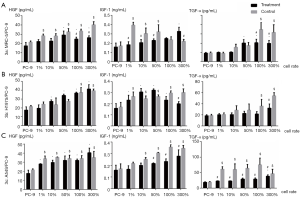
Production of HGF, IGF-1 and TGF-α in co-culture supernatant
In the three co-culture systems, whether with or without erlotinib, HGF levels in co-culture supernatants increased to different levels with the increasing proportion of cells in the upper chamber relative to PC-9 cells from 1% to 300% (P<0.05). Of note, HGF levels reached a plateau in the 1% A549/PC-9 co-culture system in both the erlotinib+ and erlotinib− subgroups. In the erlotinib− subgroup, the production of IGF-1 reached a plateau in 1% MRC5/PC-9 or H1975/PC-9 co-culture systems.
In all three co-culture systems, the production of TGF-α in the erlotinib+ subgroup was significantly lower than that in the corresponding erlotinib− subgroup, suggesting that erlotinib significantly inhibits the secretion of TGF-α in co-culture systems. In the non-erlotinib subgroup, the production of TGF-α showed a significant increase and quickly reached a plateau in the 1% A549/PC-9 co-culture system compared with PC-9 alone. For the other two co-culture systems of H1975/PC-9 or MRC-5/PC-9, TGF-α increased linearly with the increasing proportion of upper chamber H1975 or MRC-5 cells relative to lower chamber PC-9 cells (Figure 3).
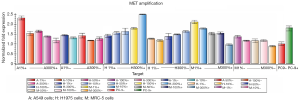
MET amplification of co-cultured PC-9 cells
MET amplification exhibited an increasing trend in the erlotinib+ subgroup of the H1975/PC-9 co-culture system with the increasing proportion of upper chamber H1975 cells. However, in the erlotinib+ subgroups of A549/PC-9 or MRC-5/PC-9 co-culture systems, MET amplification showed a decreasing trend. In the non-erlotinib subgroups of the three co-culture systems, MET amplification showed no significant changes with the increasing ratio of upper chamber cells (Figure 4).
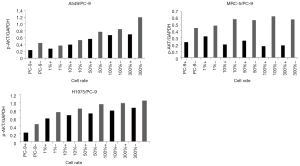
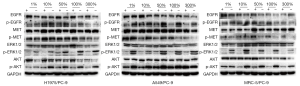
Protein expression of EGFR, MET, AKT, and ERK in co-cultured PC-9 cells
In the H1975/PC9, A549/PC-9 or MRC-5/PC-9 co-culture groups, or PC-9 alone, the proteins expressions of pEGFR, pMET, pAKT, especially pERK in the erlotinib+ subgroup, significantly decreased compared with those in the erlotinib− subgroup. The protein expression of pAKT showed a gradually ascending tendency with the increased proportion of upper chamber cells from 1% to 300% in all three co-culture systems. However, unlike the A549/PC-9 or MRC-5/PC-9 co-culture systems which showed a slow increase in pAKT protein, the H1975/PC-9 co-culture system showed a high level of pAKT increase at 1% of H1975/PC-9 compared with PC-9 alone, and rapidly reached a plateau (Figures 5-7).
Discussion
Intratumor heterogeneity provides the fuel for tumor evolution and drug resistance (15). It has been demonstrated that more than 75% of NSCLCs carry a subclonal driver alteration that occurred later in evolution (16). Soucheray and colleagues found that heterogeneity within NSCLC cell lines harboring EGFR kinase domain mutations could result in divergent resistance mechanisms in response to EGFR-TKI treatment (17). However, the interactive mechanism between the pressure imposed by EGFR-TKI treatment on tumor heterogeneity or TME and subsequent drug resistance is still poorly understood. Our findings provide evidence that tumor heterogeneity is an important reason for the resistance of lung cancer cells with EGFR-sensitive mutation to EGFR-TKIs, and suggest that the possible mechanisms may be correlated with MET amplification or AKT signaling pathways activated by paracrine of HGF, IGF-1, and TGF-α. We also found that heterogeneous EGFR T790M cells could induce MET amplification of co-cultured EGFR-sensitive mutant cells in a proportion-dependent manner when treated with erlotinib, which can explain the co-existence of secondary T790M and MET amplification in some patients after they have become resistant to EGFR-TKIs.
The development of high-through put next-generation sequencing technology has enabled great progress, and related studies have revealed extensive intratumoral heterogeneity (18). Recently, more drug-resistance mechanisms related to EGFR-TKIs have been intensively studied, but only a few linked the TME and heterogeneity together (19-21). Tumor spatial-temporal heterogeneity always changes along with EGFR-TKI treatment. It is reasonable to speculate that a pre-treatment minor subclone might emerge as a dominant resistant clone following clonal selection upon development of resistance to EGFR-TKIs. The T790M mutation or MET amplification may pre-exist in cancer cells with a small amount of cloning before targeted therapy (10), and the proportion of sensitive mutant lung cancer cells decrease after EGFR-TKI treatment, so the relative proportion of drug-resistant cells will increase; this is called clonal selection under targeted therapy, and results in the development of resistance (22,23). Ramirez and colleagues compared persister-derived erlotinib-resistant colonies derived from EGFR-addicted lung cancer cells. They then used a combination of large-scale drug screening of cells and whole-genome sequencing, and found that erlotinib-resistant colonies acquired diverse resistance mechanisms. Thus, the drug-tolerant persisted state does not limit the emergence of heterogeneous drug-resistance mechanisms (24). In our present study that linked the TME and tumor heterogeneity, tumor heterogeneity was simulated by co-culture using transwell technology to study the crosstalk between cell types with various changes. We found that the activities of PC-9 cells when treated with erlotinib increased with the increasing proportion of co-cultured heterogeneous A549 cells or MRC-5 cells. Of note, no change in activity of PC-9 cells was observed in the H1975/PC-9 co-culture system. These findings indicate that both heterogeneous tumor cells and stromal cells in the TME could influence the sensitivity of EGFR-sensitive mutant cells to EGFR-TKIs, suggesting that crosstalk among heterogeneous tumor cells and stromal cells during EGFR-TKI treatment may influence tumor sensitivity to EGFR-TKIs and be correlated with primary resistance. However, not all heterogeneous tumor types in the TME, for example preexisting T790M mutant cells, influence the sensitivity to EGFR-TKIs by a direct crosstalk mechanism. Hata and colleagues have recently demonstrated, using patient-derived cell line models, that T790M drug-resistant NSCLC cells can both preexist and evolve from drug-tolerant cells (25). This partly explains why some patients without pre-treatment T790M developed acquired resistance caused by the T790M mutation. The authors simultaneously found that those T790M drug-resistant cells that evolved from drug-tolerant cells had a diminished apoptotic response to third-generation EGFR-TKI.
The mechanism of EGFR-TKI resistance caused by tumor heterogeneity was further investigated in our studies. We found that in cases of heterogeneity, MET amplification may not be the main mechanism responsible for rendering PC-9 cells unresponsive to EGFR-TKI, because decreased MET amplification was observed in PC-9 cells with increased viability in A549/PC-9 and MRC-5 co-culture systems, while increased MET amplification was observed in PC-9 cells without altered viability in the H1975/PC-9 co-culture system. We found that in all three co-culture systems, the expression of pAKT tended to increase with the increasing proportion of upper chamber cells from 1% to 300%, suggesting that bypass of AKT activation caused by tumor heterogeneity might be involved in primary resistance to EGFR-TKIs. Our study results were in accord with a study conducted by Suda et al., who found that resistant wild-type lung cancer cells induced by erlotinib treatment could express much more pAKT and pERK1/2 proteins than those original wild type lung cancer cells without erlotinib treatment (26).
What drives the activation of the AKT signaling pathway is a key issue. It has been proven that HGF plays a very important role in drug resistance. HGF was found to be overexpressed in 30% of NSCLC patients with an EGFR-sensitive mutation who had primary resistance to EGFR-TKIs (27). HGF overexpression has been found more frequently than T790M mutation or MET amplification in acquired drug resistance (28). The result of our current study showed that the HGF secreted in the three co-culture systems showed a similar increase with the increase in the proportion of the upper cells regardless of erlotinib treatment, but no difference was found between the erlotinib+ and erlotinib− subgroups. There is still insufficient evidence to conclude that HGF is associated with primary or secondary drug resistance in tumor heterogeneity. However, there is more and more evidence that the IGF-1 receptor pathway could mediate acquired resistance to EGFR-TKI. IGF-1 is secreted by tumor cells through autocrine or paracrine mechanisms. After IGF-1 binds to its receptor, the PI3K/AKT and RAS/RAF/MEK/ERK signal pathways can be activated, thus promoting tumor cell proliferation, and suppressing apoptosis (29-31). Our in vitro experiment found that IGF-1 gradually increased with increasing numbers of A549 cells in both the erlotinib+ and the erlotinib− groups, indicating that more wild-type lung cancer cells in the TME may induce more IGF-1 secretion. A large amount of IGF-1 could thus be secreted by a small proportion of H1975 or MRC-5 cells in a PC-9 co-culture system without erlotinib treatment. However, the opposite trend of change was observed in the erlotinib treatment subgroup. This may mean the more obvious the intertumoral heterogeneity, the more complex the cytokines secreted. It is difficult to explain the reason for this in the present study, and further studies are needed to confirm this phenomenon. Of note, a small proportion of A549 or MRC-9 cells co-cultured with PC-9 produced large amounts of TGF-α in the erlotinib− subgroups, but after erlotinib treatment the TGF-α level significantly decreased. This suggests that TGF-α may be, to some extent, related to primary drug resistance. Previous studies have shown that TGF-α, like epidermal growth factor, can phosphorylate the ATP binding site of tyrosine kinases by binding with EGFR, activating EGFR downstream signal transduction, and resulting in drug resistance of EGFR-sensitive mutant NSCLCs (32). There have been studies verifying that antibodies against HGF, TGF-α and IGF-1 could inhibit the growth of EGFR-sensitive mutant lung cancer cells (26,32-34). Thus, we believe that the HGF, IGF-1 and TGF-α produced via paracrine secretion by heterogeneous cells, even under the influence of EGFR-TKIs, could activate the downstream signaling pathways of EGFR, especially PI3K-AKT, thus inducing resistance to EGFR-TKIs. This may be one of the important mechanisms linking drug-resistance and TME and tumor heterogeneity imposed by EGFR-TKI selection pressure. These findings suggest that receptor ligands of HGF, TGF-α and IGF-1 secreted by TME may be additional targets during treatment.
There are some limitations to this work. The effect of the microenvironment and tumor heterogeneity on drug resistance of lung cancer is very important but complicated. There are many other factors which influence the TME such as oxygen content, drugs, or glucose levels. The established co-culture models only simulated crosstalk of pure cell to cell and do not account for many factors present in the microenvironment. As shown in many studies, cancer cells are regulated by much more than one cytokine or one mesenchymal cell. Of course, there were many cytokines that we did not detect, such as TGF-β1, platelet derived growth factorα/β (PDGF-α/β), basic fibroblast growth factor (b-FGF), or interleukin 6 (IL-6) (35). Therefore, there are still many unknown factors which need to be explored. In addition, the TME changes dynamically and complexly. Researchers used a novel experimental model derived from the HCC827 erlotinib-sensitive and -resistant NSCLC cell lines. They found that in contrast to in the drug-free environment, resistant cells may hold some advantage compared to parental cells in microenvironments deficient in oxygen and nutrients (20). TME compounded by variable cell density, differential forces on cells, effect of these forces on growth rates, and consumption of nutrients by cells (20). Furthermore, we did not analyze and compare the differences at different time-points among these co-culture systems. Thus a complex and approximate real microenvironment model will be the topic of future investigation.
Conclusions
Our in vitro gradient co-culture experiment indicated that heterogeneous lung cancer cells and fibroblasts could produce large amounts of HGF, TGF-α and IGF-1, especially when these cells were present in greater proportions. Tumor heterogeneity is an important reason for the resistance of lung cancer cells with an EGFR-sensitive mutation to EGFR-TKI, and one possible mechanism may be related to activation of the downstream signaling pathways of EGFR-AKT by paracrine secretion of HGF, TGFα and IGF-1. MET amplification of EGFR-sensitive mutant cells presented an increasing trend with the increasing proportion of heterogeneous EGFR T790M cells, helping to explain the co-existence of secondary T790M and MET amplification after development of resistance to EGFR-TKIs.
Acknowledgments
Funding: This study was supported by grants from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was based on the experiments conducted in vitro using the cell line available for purchase. Ethical approval and individual informed was not required.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol 2016;893:1-19. [Crossref] [PubMed]
- NSCLC Meta-Analyses Collaborative Group. Chemotherapy in addition to supportive care improves survival in advanced nonsmall-cell lung cancer: a systematic review and meta-analysis of individual patient data from 16 randomized controlled trials. J Clin Oncol 2008;26:4617-25. [Crossref] [PubMed]
- Okabe T, Okamoto I, Tamura K, et al. Differential constitutive activation of the epidermal growth factor receptor in non-small cell lung cancer cells bearing EGFR gene mutation and amplification. Cancer Res 2007;67:2046-53. [Crossref] [PubMed]
- Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol 2011;29:2866-74. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [Crossref] [PubMed]
- Yang JC, Schuler MH, Yamamoto N, et al. LUX-Lung 3: A randomized, open-label, phase III study of afatinib versus pemetrexed and cisplatin as first-line treatment for patients with advanced adenocarcinoma of the lung harboring EGFR-activating mutations. J Clin Oncol 2017;30:LBA7500. [Crossref]
- Zhong WZ, Zhou Q, Wu YL. The resistance mechanisms and treatment strategies for EGFR-mutant advanced non-small-cell lung cancer. Oncotarget 2017;8:71358-70. [Crossref] [PubMed]
- Shien K, Yamamoto H, Soh J, et al. Drug resistance to EGFR tyrosine kinase inhibitors for non-small cell lung cancer. Acta Med Okayama 2014;68:191-200. [PubMed]
- Cheng X, Chen H. Tumor heterogeneity and resistance to EGFR-targeted therapy in advanced non-small cell lung cancer: challenges and perspectives. Onco Targets Ther 2014;7:1689-704. [Crossref] [PubMed]
- Chen ZY, Zhong WZ, Zhang XC, et al. EGFR mutation heterogeneity and mixed response to EGFR tyrosine kinase inhibitors of nonsmall cell lung cancer: a clue to overcoming resistance. Oncologist 2012;17:978-85. [Crossref] [PubMed]
- Bai H, Wang Z, Wang Y, et al. Detection and Clinical Significance of Intratumoral EGFR Mutational Heterogeneity in Chinese Patients with Advanced Non-Small Cell Lung Cancer. PLoS One 2013;8:e54170. [Crossref] [PubMed]
- Zhou Q, Zhang XC, Chen ZH, et al. Relative Abundance of EGFR Mutations Predicts Benefit from Gefitinib Treatment for Advanced Non–Small-Cell Lung Cancer. J Clin Oncol 2011;29:3316-21. [Crossref] [PubMed]
- Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A 2007;104:20932-7. [Crossref] [PubMed]
- Greaves M. Evolutionary determinants of cancer. Cancer Discov 2015;5:806-20. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med. 2017;376:2109-21. [Crossref] [PubMed]
- Soucheray M, Capelletti M, Pulido I, et al. Intratumoral Heterogeneity in EGFR-Mutant NSCLC Results in Divergent Resistance Mechanisms in Response to EGFR Tyrosine Kinase Inhibition. Cancer Res 2015;75:4372-83. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer 2017;17:637-658. [Crossref] [PubMed]
- Mumenthaler SM, Foo J, Choi NC, et al. The Impact of Microenvironmental Heterogeneity on the Evolution of Drug Resistance in Cancer Cells. Cancer Inform 2015;14:19-31. [PubMed]
- Whiteside TL. Exosome and mesenchymal stem cell cross-talk in the tumor microenvironment. Semin Immunol 2018;35:69-79. [Crossref] [PubMed]
- Foo J, Michor F. Evolution of resistance to anti-cancer therapy during general dosing schedules. J Theor Biol 2010;263:179-88. [Crossref] [PubMed]
- Sun D, Lauffenburger DA, Zhao B, et al. Differential selective pressure alters rate of drug resistance acquisition in heterogeneous tumor populations. Sci Rep 2016;6:36198. [Crossref] [PubMed]
- Ramirez M, Rajaram S, Steininger RJ, et al. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat Commun 2016;7:10690. [Crossref] [PubMed]
- Hata AN, Niederst MJ, Archibald HL, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med 2016;22:262-9. [Crossref] [PubMed]
- Suda K, Mizuuchi H, Sato K, et al. The insulin-like growth factor 1 receptor causes acquired resistance to erlotinib in lung cancer cells with the wild-type epidermal growth factor receptor. Int J Cancer 2014;135:1002-6. [Crossref] [PubMed]
- Song S, Bi M. Research progress of HGF/MET signaling pathway in EGFR-TKI resistance in non-small cell lung cancer. Zhongguo Fei Ai Za Zhi 2014;17:755-9. [PubMed]
- Yano S, Takeuchi S, Nakagawa T, et al. Ligand-triggered resistance to molecular targeted drugs in lung cancer: roles of hepatocyte growth factor and epidermal growth factor receptor ligands. Cancer Sci 2012;103:1189-94. [Crossref] [PubMed]
- Shelton JG, Steelman LS, White ER, et al. Synergy between PI3K/Akt and Raf/MEK/ERK pathways in IGF-1R mediated cell cycle progression and prevention of apoptosis in hematopoietic cells. Cell Cycle 2004;3:372-9. [PubMed]
- Morinaga R, Okamoto I, Furuta K, et al. Sequential occurrence of non-small cell and small cell lung cancer with the same EGFR mutation. Lung Cancer 2007;58:411-3. [Crossref] [PubMed]
- Wang J, Wang B, Chu H, et al. Intrinsic resistance to EGFR tyrosine kinase inhibitors in advanced non-small-cell lung cancer with activating EGFR mutations. Onco Targets Ther 2016;9:3711-26. [Crossref] [PubMed]
- McClintock JL, Ceresa BP. Transforming growth factor-{alpha} enhances corneal epithelial cell migration by promoting EGFR recycling. Invest Ophthalmol Vis Sci 2010;51:3455-61. [Crossref] [PubMed]
- Yano S, Wang W, Li Q, et al. Hepatocyte Growth Factor Induces Gefitinib Resistance of Lung Adenocarcinoma with Epidermal Growth Factor Receptor-Activating Mutations. Cancer Res 2008;68:9479-87. [Crossref] [PubMed]
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43. [Crossref] [PubMed]
- Cirri P, Chiarugi P. Cancer-associated-fibroblasts and tumour cells: a diabolic liaison driving cancer progression. Cancer Metastasis Rev 2012;31:195-208. [Crossref] [PubMed]