
N6-methyladenosine could indirectly modulate translation in human cancer cells via cis-elements
Introduction
N6-methyladenosine (m6A) is one of the common forms of RNA modifications, which is found to occur in RNAs of many organisms or cell lines (1-9). The distribution of m6A on mRNAs are not random, it is reported that the methylated adenosine is located in GRAC, RRACH or DRACH motif (R = A or G, H = A, C or T, D = A, G or T). In mammals, one out of one thousand adenosines in total RNAs are N6-methylated (5,6,8,10). Over ten thousand m6A modification sites have been identified in mRNAs (4-6,8,11,12), which are enriched in long exons, 3'UTRs and around stop codons. The m6A methylation events are being regulated by different factors and meanwhile regulating the downstream biological processes. The dynamic regulation of m6A modification is accomplished by the m6A writers (methyltransferase complex) (13,14) and erasers (15,16), and the downstream effects of m6A are exerted with the help of readers (5,6,11,17) or by m6A itself (1,4).
The writer complex is mainly composed of METTL3, WTAP and METTL14. METTL3 is the core catalytic enzyme and METTL14 acts as a structural role (18). WTAP is found to stabilize the dimerized METTL3 and METTL14. Other proteins like RBM15 and KIAA1429 were also found in the writer complex (19). Apart from the preference in RRACH motifs, based on the fact that the methylation process is co-transcriptional (20), recent studies revealed that METTL14 interacts with histone modifications and promote site-specific methylation on pre-mRNAs (21). The reversibility of methylation indicates the presence of eraser proteins. Two erasers FTO and ALKBH5 are found in mammals. The lack of FTO leads to abnormal growth, development, mobility and metabolism (22-24). The mutated ALKBH5 would affect spermatogenesis, and also cause cancer (25,26).
The reader proteins of m6A modification are important for executing the biological functions of m6A. In mammals, m6A readers mainly include YTHDF1/2/3 and YTHDC1/2. Phenotypical and functional studies discovered that the readers are important for stem cell development and cancer immunity (27-29). Meanwhile, the mechanistic studies revealed profound molecular details of how m6A sites interact with reader proteins. Binding of YTHDF2 on m6A transcripts affects the stability of the mRNAs (5), and YTHDF1 recruits the translation initiation factors (particularly eIF3) to m6A modified mRNAs to enhance the translation efficiency (TE) of these genes (6). Meanwhile, m6A is also able to modulate mRNA translation independent of reader proteins. Under heat stress, m6A on 5'UTR of Hsp70 mRNAs acts as IRES (internal ribosome entrance site) to facilitate the translation initiation (by eIF3) of Hsp70 (9,30).
The cases of YTHDF1 (6) and IRES (9,30) are both related to the initiation factor(s) eIF3 that could directly determine the translation initiation. We wonder whether m6A could fine-tune the translation process through other indirect approaches. Interestingly, it is reported that m6A modification could alter the splicing patterns of mRNAs and this change even affects the sex determination in Drosophila (1,3,4). This also indicates the case that m6A modification could exert its function without the need of reader proteins. This phenomenon reminds us that m6A might be able to modulate translation through other indirect ways. It is established that many cis-elements in the 5'UTR are important for determining the TE of downstream CDS, such as sequence motifs for scanning, structures and non-canonical reading frames. If the splicing changes caused by m6A could lead to the gain or loss of these decisive cis-elements in 5'UTRs, this would eventually affect the translation of host genes.
To test our hypothesis, we retrieved the m6A genes in HeLa cells. In the METTL3 knock-down libraries (6), we examined the global changes in mRNA splicing patterns as well as TE. The differential splicing (DS) genes are enriched in m6A modified genes and the DS events are relatively enriched in 5'UTRs. The 105 genes with DS events in 5'UTR alter their TE more strongly than the genes with DS events in other regions (CDS/3'UTR/intron). Furthermore, the splicing pattern of 98 out of those 105 genes are unaffected by reader YTHDF1. Importantly, we did not observe significant TE changes for these 98 genes when YTHDF1 was knocked down.
Our results demonstrate that m6A could modulate the translation of mRNAs through affecting the splicing patterns, at least for a small set of genes. These indirect effects are independent of the direct regulation by reader proteins. Our work extended our knowledge about the translation regulation by m6A.
Methods
Next generation sequencing (NGS) data
The NGS (next generation sequencing) data in normal HeLa cells or HeLa cells with si-METTL3/si-YTHDF1 were downloaded from a previous study (6). The NGS data contain mRNA-Seq and Ribo-Seq (ribosome profiling followed by deep sequencing) (31) that enable us to define the TE of each gene. Those adenosine sites in GRAC motif (if located in m6A peaks) were systematically recognized as m6A modification sites. Note that throughout this manuscript, the “si-” and “-KD” mean the same. They are used to avoid monotonic expressions.
Assigning the m6A sites to the human genes
We downloaded the hg19 human genome (UCSC). In determining whether an m6A site is located in the 5'UTR/CDS/3'UTR/intron of a gene, we chose the longest isoform of each gene, however in cases where isoforms had the same length, they were sorted alphabetically. This way, each m6A modification site has a unique gene ID, transcript (isoform) ID and functional category (5'UTR/CDS/3'UTR/intron or noncoding).
NGS data processing
Following the previous study, the sequencing reads were aligned to human genome (hg19) with STAR (32). The default parameters were used. The read counts for each gene were counted using htseq-count (33) with default parameters. Only uniquely mapped reads were counted. The “expressed genes in HeLa cells” are defined as those genes with raw read count >50 in si-control cells.
Differential splicing (DS) genes
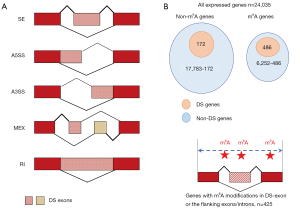
We used rMATS (34) to determine the DS genes in si-METTL3 or si-YTHDF1 vs. control. Note that mRNA-Seq rather than Ribo-Seq should be used for DS analysis. In the output file of rMATS software, if a splicing event has false discovery rate (FDR) <0.05 (35), it is recognized as a DS event, and the “host gene” of this DS event is termed DS gene. Accordingly, the “host exon” that contains the DS event is termed DS exon. There are five types of splicing according to the rMATS manual (Figure 1A). Any types of splicing with FDR <0.05 are regarded as DS events. When calculating the enrichment of DS events, since m6A modifications are enriched in 3'UTRs and around stop codons, we should not directly count the number of DS events caused by si-METTL3. We took the m6A distribution into consideration (as background). Take CDS as an example, we define (I) percentage of m6A sites in CDS = number of m6A sites in CDS/total number of m6A sites on mRNA; (II) percentage of DS events in CDS = number of DS events in CDS/total number of DS events; (III) enrichment of DS events in CDS = percentage of DS events in CDS/percentage of m6A sites in CDS.
TE of genes
We utilized TE = RPKM in Ribo-Seq/RPKM in mRNA-Seq (6,9,30) to determine the TE of each gene or the TE-foldchange between si-METTL3 or si-YTHDF1 vs. control libraries. RPKM stands for reads per kilobase per million (reads). Note that when calculating TE, only the reads mapped to CDS regions are counted.
Statistical analyses
We used R language to conduct statistical analyses (https://www.R-project.org/). The comparisons of TE-foldchange values were performed using Wilcoxon rank sum tests. The enrichment comparison (between fractions) was performed using Fisher’s exact tests. The statistical significance was denoted as: *, P<0.05; **, P<0.01; ***, P<0.001.
Results
Splicing changes when METTL3 was knocked down were enriched around m6A exons
There are five types of alternative splicing patterns, SE (skipped exon), A5SS (alternative 5' splice site), A3SS (alternative 3' splice site), MEX (multiple exclusive exon) and RI (retained intron) (Figure 1A). In the mRNA-Seq data of si-METTL3 vs. control, any types of splicing with FDR <0.05 are defined as DS events (Methods). The genes with DS events are defined as DS genes when METTL3 was knocked down.
Among the 24,035 expressed human genes in HeLa cells, 6,252 are m6A genes and 17,783 are non-m6A genes (Figure 1B). Interestingly, 486/6,252 (7.8%) out of the m6A genes and 172/17,783 (0.97%) out of the non-m6A genes are DS genes when METTL3 was knocked down (P value <0.001, Fisher’s exact test; Figure 1B). Furthermore, we found that 425/486 (87.4%) of the DS genes had m6A modifications on the DS exon or the on the exons/introns flanking the DS exon (Figure 1B), indicating that the m6A sites close to the DS events might be responsible for the splicing changes.
DS events are relatively enriched in 5'UTRs
We questioned whether the DS events have preference in mRNA locations. We focus on the DS events in the 486 DS m6A genes and calculated the relative enrichment of DS events in each mRNA location (Methods). We found that the enrichment of DS events in 5'UTR is greater than 1 while those of other categories (CDS, 3'UTR, intron or noncoding) do not significantly differ from 1 (Figure 2A). Our next question is based on the fact that many cis-elements in 5'UTR of mRNA play essential roles in determining the translation initiation, so could the DS events in 5'UTR cause the gain or loss of these essential cis-elements and consequently affect the translation of host genes?
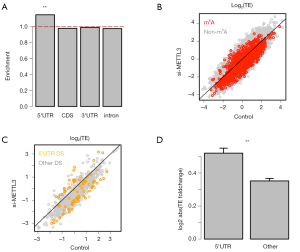
DS events in 5'UTRs cause stronger alteration of TE
We profiled the TE foldchange of all genes in si-METTL3 vs. control. The TE of the 6,252 m6A genes are globally down-regulated when METTL3 was knocked down (Figure 2B), which agrees with previous knowledge (6). Among the 486 DS m6A genes, the genes with DS events in 5'UTR are highlighted against genes with DS events in other regions (CDS, 3'UTR and intron are combined) (Figure 2C). We could see that the gene with DS events in 5'UTR tend to have greater changes in TE (either increase or decrease). Furthermore, the absolute values of TE foldchange clearly show that the genes with DS events in 5'UTR alter their TE more severely than those with DS events in other regions (Figure 2D).
Translation of the 5'UTR DS genes is generally unaffected by YTHDF1
For the 425 m6A genes with DS events near m6A sites (Figures 1B,3), 105 have DS events in 5'UTR (Figure 3). Our hypothesis is that the translation of these 5'UTR DS genes might be affected by additional cis elements, not only the reader protein alone (this does not mean the readers do not contribute). First, we should confirm that these 105 genes are not differentially spliced in YTHDF1-KD. 98 out of the 105 genes are non-DS genes in YTHDF1-KD (Figure 3), suggesting that the splicing changes observed in METTL3-KD are mainly caused by m6A modifications rather than reader proteins.
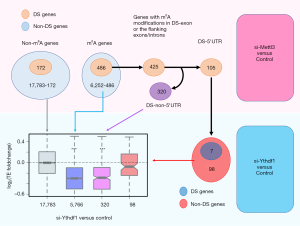
We next investigated the TE foldchange in YTHDF1-KD vs. control. Different gene sets showed distinct patterns (Figure 3). For the 17,783 non-m6A genes (background), their TE log2-foldchanges had a median value around zero, which agreed with our expectation. For the 5,766 non-DS m6A genes (in METTL3-KD), their TEs were significantly down-regulated in YTHDF1-KD (Figure 3). The same trend of decreased TE went for the 320 DS m6A genes which had DS events in non-5'UTR regions (Figure 3). This indicated that m6A genes would be translationally down-regulated when reader protein is knocked down. For the 98 DS genes in 5'UTR, their TE was only slightly and insignificantly down-regulated in YTHDF1-KD (Figure 3). This result proves that the TE changes of the 98 [105] genes observed in METTL3-KD were majorly contributed by DS-related cis changes. The reader protein alone could not account for such a big change (Figure 2C,D) as demonstrated in Figure 3.
Discussion
RNA modifications like the m6A methylation is highly regulated by different factors and meanwhile regulating its downstream biological processes. As we have introduced, the dynamic regulation of m6A modification is accomplished by the writers (13,14) and erasers (15,16), and the downstream effects of m6A are exerted with the help of readers (5,6,11,17,36) or by m6A site itself (1,4). However, it is not strange at all that the effects of “readers” and “m6A site itself” could simultaneously contribute to the translation status of host genes. Our goal is to search for the minor and indirect effects of m6A site itself on the translation of host genes.
By comparing the global changes in mRNA splicing patterns and TE of si-METTL3 vs. control in human HeLa cells, we found that the DS genes are enriched in m6A modified genes, suggesting that the DS events (in si-METTL3 vs. control) are likely caused by m6A modifications. Changes in splicing patterns are often related to 5'UTR. The global TE of m6A modified genes (no matter the modification is on UTR or CDS) is down-regulated when METTL3 was knocked down. This result agrees with previous knowledge that m6A enhances the translation of host gene by the reader protein (6). We further found that the genes with DS events in 5'UTR alter their TE more severely than those genes with DS events in CDS/3'UTR/intron, suggesting a role of m6A-mediated splicing changes in translation regulation. Importantly, we verified our hypothesis in the YTHDF1-KD samples. The TE of the few 5'UTR DS genes in METTL3-KD was unchanged in YTHDF1-KD (Figure 3), indicating that their TE changes in METTL3-KD might be largely due to the effect of DS caused by m6A itself.
The established theory of how m6A affects mRNA translation includes (I) YTHDF1 recruits the initiation factors to m6A modified mRNAs and promote the translation (6) and (II) m6A on 5'UTR acts as IRES to directly initiate translation (9,30). In contrast to these big findings by previous literatures, our analysis suggests the fine-tuning of mRNA translation mediated by m6A modification and splicing changes. These fine-tuning effects are indirect and relatively weaker than those direct approaches.
One might think that “independent of reader proteins” could be a new mechanism. However, we do not wish to overstate this conclusion. It only account for a few genes that have 5'UTR m6A sites and meanwhile have DS events in 5'UTR when m6A is knocked down (conservatively 98 genes). Let us take the “microRNA binding to 3'UTR” as an example. Presume that an m6A site is located in 3'UTR of a gene and the suppression of methylation results in DS in 3'UTR region. If this DS event in 3'UTR leads to the gain or loss of microRNA binding sites, then it might eventually cause changes in translation or mRNA expression of host gene. We think this example could not be called a new mechanism because (I) “m6A affects splicing” is known; (II) “Splicing might cause gain or loss of microRNA binding site” is conceivable; (III) “microRNA binding affects host gene translation or expression” is known. There are only a few candidate genes that could fit all these criteria. This pattern is merely valid for specific genes rather than all genes (just because these few gene happened to fit the criteria). Similarly, our “m6A-splicing-5'UTR-translation” chain is based on known mechanisms in every step. Only a small set of genes could link all these steps together (conservatively 98 genes in Figure 3). Our purpose is to reveal this observation and show that the m6A modification could affect the host gene through this indirect way.
Next, we want to clarify that our results do not conflict with known theories. The translational effect caused by 5'UTR DS events is “independent of reader proteins” does not mean that the reader proteins contribute nothing. We should clearly clarify that “both the 5'UTR DS events and the reader protein contribute to the observed translational changes”. The YTHDF1-dependent pathway to enhance the translation of m6A genes was discovered years ago (6). We think that the translational changes are caused by multiple factors. It is like the multiple regression analysis. The global trend showed that the TE of m6A genes were down-regulated in YTHDF1-KD (6), which proved the positive contribution of YTHDF1. For some m6A genes with up-regulated TE, the elevated TE might be explained by other variables (like the gain or loss of some cis-elements in UTRs). Similarly, in our analyses, we found 98 genes that their DS events strongly affects their translation and the reader protein YTHDF1 plays a less important role. This does not mean that YTHDF1 does not contribute to the translation of target genes. Thus, our findings do not conflict with known theories.
Our work might remind people that the translation status is affected by numerous factors and none of the factors alone could explain all the observed translational changes. Although the global comparison showed that thousands of m6A genes were translationally down-regulated when YTHDF1 was knocked down (6), one could always find the exceptions that a few m6A genes were up-regulated, which might be caused by other factors including cis elements in UTRs. At this stage, the detailed mechanism of how DS events affect the translation of host gene remains unexplored. What we know is that there are many cis-elements in 5'UTR that determine the translation initiation, and that the DS events in 5'UTR might cause the gain or loss of these important cis-elements and eventually impact the translation of downstream CDS.
Conclusions
Our results reveal that m6A modifications could modulate the translation of mRNAs through affecting their splicing patterns. The differential splicing caused by m6A are enriched in 5'UTR and consequently affect translation. These indirect effects are independent of the direct regulation by reader proteins, and likely to be related to the cis-elements in 5'UTR that determine the translation of downstream CDS. Our work broadened our knowledge about the translation regulation by m6A modifications.
Acknowledgments
We thank the members in our Hospital that have given suggestions to our project.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.18). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and individual informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Haussmann IU, Bodi Z, Sanchez-Moran E, et al. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 2016;540:301-4. [Crossref] [PubMed]
- Huang H, Weng H, Sun W, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 2018;20:285-95. [Crossref] [PubMed]
- Kan L, Grozhik AV, Vedanayagam J, et al. The m(6)A pathway facilitates sex determination in Drosophila. Nat Commun 2017;8:15737. [Crossref] [PubMed]
- Lence T, Akhtar J, Bayer M, et al. m(6)A modulates neuronal functions and sex determination in Drosophila. Nature 2016;540:242-7. [Crossref] [PubMed]
- Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014;505:117-20. [Crossref] [PubMed]
- Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015;161:1388-99. [Crossref] [PubMed]
- Xu K, Yang Y, Feng GH, et al. Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res 2017;27:1100-14. [Crossref] [PubMed]
- Zheng Y, Nie P, Peng D, et al. m6AVar: a database of functional variants involved in m6A modification. Nucleic Acids Res 2018;46:D139-45. [Crossref] [PubMed]
- Zhou J, Wan J, Gao X, et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015;526:591-4. [Crossref] [PubMed]
- Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 2017;18:31-42. [Crossref] [PubMed]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012;485:201-6. [Crossref] [PubMed]
- Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 2012;149:1635-46. [Crossref] [PubMed]
- Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 2014;10:93-5. [Crossref] [PubMed]
- Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017;169:1187-200. [Crossref] [PubMed]
- Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011;7:885-7. [Crossref] [PubMed]
- Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013;49:18-29. [Crossref] [PubMed]
- Xiao W, Adhikari S, Dahal U, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell 2016;61:507-19. [Crossref] [PubMed]
- Huang J, Yin P. Structural Insights into N6-methyladenosine (m6A) Modification in the Transcriptome. Genomics Genomics Proteomics Bioinformatics 2018;16:85-98. [Crossref] [PubMed]
- Patil DP, Chen CK, Pickering BF, et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016;537:369. [Crossref] [PubMed]
- Ping XL. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 2014;24:177-89. [Crossref] [PubMed]
- Huang H, Weng H, Zhou K, et al. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 2019;567:414-9. [Crossref] [PubMed]
- Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet 2007;39:724-6. [Crossref] [PubMed]
- Fischer J, Koch L, Emmerling C, et al. Inactivation of the Fto gene protects from obesity. Nature 2009;458:894-8. [Crossref] [PubMed]
- Boissel S, Reish O, Proulx K, et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am J Hum Genet 2009;85:106-11. [Crossref] [PubMed]
- Cui Q, Shi H, Ye P, et al. m(6)A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep 2017;18:2622-34. [Crossref] [PubMed]
- Zhang C, Zhi WI, Lu H, et al. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget 2016;7:64527-42. [PubMed]
- Zhang C, Chen Y, Sun B, et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature 2017;549:273. [Crossref] [PubMed]
- Barbieri I, Tzelepis K, Pandolfini L, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 2017;552:126. [Crossref] [PubMed]
- Han D, Liu J, Chen C, et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature 2019;566:270-4. [Crossref] [PubMed]
- Meyer KD, Patil DP, Zhou J, et al. 5 ' UTR m(6)A Promotes Cap-Independent Translation. Cell 2015;163:999-1010. [Crossref] [PubMed]
- Ingolia NT, Ghaemmaghami S, Newman JR, et al. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009;324:218-23. [Crossref] [PubMed]
- Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15-21. [Crossref] [PubMed]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166-9. [Crossref] [PubMed]
- Shen S, Park JW, Lu ZX, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A 2014;111:E5593-601. [Crossref] [PubMed]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological 1995;57:289-300. [Crossref]
- Chu D, Wei L. Human cancer cells compensate the genes unfavorable for translation by N6-methyladenosine modification and enhance their translation efficiency. Transl Cancer Res 2019;8:499-508. [Crossref]


