
MicroRNA let-7a inhibits proliferation of breast cancer cell by downregulating USP32 expression
Introduction
MicroRNAs (miRs) are a class of non-coding RNA molecules that function as regulators of target gene. They inhibit target proteins expression through inducing the messenger RNA (mRNA) degradation or repressing translation (1-3). miRs are known to be aberrantly expressed as tumor suppressors or oncogenes in cancer, and closely related to the physiological cellular processes such as cell cycle, cell proliferation and cell apoptosis (4-6). miR let-7a, as a tumor suppressor, is a member of miR let-7 family. Several studies have demonstrated that miR let-7a inhibits cell growth by down-regulating HMGA-2 in non-small cell lung cancer (7). In gastric cancer, miR let-7a targets PKM2 to inhibit cell proliferation (8). In nasopharyngeal carcinoma, miR let-7a inhibits cell proliferation by targeting EZH2 (9). In renal cell carcinoma cell lines, miR let-7a inhibits cell proliferation through targeting c-Myc pathway (10). These studies have proved that miR let-7a plays important role in cell proliferation.
Ubiquitylation is an important process of posttranslational modification with ubiquitin, and involved in almost every aspects of cell biology through regulating many processes including protein activity, cell signaling, intracellular trafficking and the DNA damage response (11). Similar to other posttranslational modifications, ubiquitination of a number of proteins can be reversed by deubiquitinases (DUBs) (12). Approximately one hundred DUBs have been identified in the mammalian genome (13), of which the ubiquitin-specific proteases (USPs) represents the largest subgroup with over 60 members, and being known to participate in cancer progression (14), DNA damage responses (15), virus infection and immune regulation (16,17). USP7 regulates Hippo signaling to modulate cell proliferation and apoptosis through deubiquitinating transcription coactivator Yorkie (18). USP10 is a tumor suppressor gene involved in the initiation of hepatocellular carcinoma and non-small cell lung cancer (19,20). USP47 and USP7 regulates inflammasome activation and the release of the proinflammatory cytokines in macrophages (21). Although USPs play a crucial role in a variety of cellular processes, the molecular mechanisms of USPs regulation remain poorly understood.
In this study, we detected the impact of miR let-7a on the protein expression of USP32 and identified USP32 as a target gene of miR let-7a. Furthermore, we investigated whether miR let-7a affected breast cancer (BCa) MCF-7 cells growth by downregulating target gene USP32. The results may provide support for the utilization of miR let-7a as novel therapeutics through targeting USP32 in BCa.
Methods
Antibodies and reagents
Rabbit polyclonal anti-USP32 (ab86792) was purchased from Abcam (Abcam, Cambridge, MA, USA). Mouse against β–actin (sc-1616) were from Santa Cruz (Santa Cruz Biotechnology, CA, USA). Their respective horseradish peroxidase-labeled polyclonal secondary antibodies were purchased from Zhongshan Golden Bridge (1:10,000, Zhongshan Golden Bridge Biological Technology, Beijing, China).
Human miR let-7a mimics, normal control (NC) mimics, miR let-7a inhibitors, NC inhibitors were synthesized by RiBoBio (Guangzhou, China). The primers used for miR let-7a and U6 were purchased from GenePharma (Shanghai, China).
Cell culture
The human BCa cells MCF-7, BT474, MDA-MB-231, HCC1937, T47D were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). MCF-7 was maintained in DME medium and BT474, MDA-MB-231, HCC1937, T47D in RPMI-1640 medium. All the medium were supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 µg/mL streptomycin. The cell lines were grown at 37 °C in a humidified incubator with 95% air and 5% carbon dioxide atmosphere.
33 Plasmid construction
USP32 full-length coding sequences were cloned into a Myc tagged CMV10 vector (Sigma) using a polymerase chain reaction (PCR) kit (Takara Biotechnology, Dalian, China). The PCR primers used to amplify the USP32 full-length coding sequences are: USP32 forward, 5'-TGCTCTAGAGGGAGCATGGGTGCCAA-3' and USP32 reverse, 5'-GCTCTAGATTACTGTAACACACAGTACTTTTTGTAATC-3'.
The wild-type (WT) 3'-untranslated region (UTR) of the human USP32 gene was amplified and cloned into the 3' end of the pMIR-REPORT Luciferase vector (Ambion Life Technologies, Carlsbad, CA, USA) between the SacI and Hind III sites, and named pMIR-USP32-3'UTRw. The USP32-3'UTR primers sequences are as follows: USP32-3'UTR forward, 5'-CGAGCTCGCTACCACTCTGGCTGCT-3' and USP32-3'UTR reverse, 5'-CCCAAGCTT GTCAGTGCATTGTCAACAAGT-3'.
Deletion mutant of the potential target site for miR let-7a located in the USP32WT-3'UTR was constructed using a two-step PCR method. Using pMIR-USP32-3'UTRw as the template, a 5' flanking region was subcloned with the primers, USP32-3'UTR forward and mutant reverse (5'-CAGGAAAAAG CATATCATGTCGTCCCT-3'), and the 3' flanking region was subcloned with the primers, USP32-3'UTR reverse and mutant forward (5'-ACATGATATGCTTTTTCCTGAAGTT-3'). The two fragments were linked with PCR, and then were cloned into the Sac I and Hind III sites of pMIR-REPORT Luciferase, and named pMIR-USP32-3'UTRm. The sequences of all the constructing plasmids were detected by sequencing.
Transfection of miRNAs and plasmids
The miR let-7a mimics, NC mimics, miR let-7a inhibitors or NC inhibitors were transfected into the MCF-7 cells at 30% confluency using Lipofectamine 2000 transfection reagent (Invitrogen) in accordance with the protocol of the manufacturer.
The pMIR-USP32-3'UTRw, pMIR-USP32-3'UTRm and pMIR-REPORT plasmids were transfected into the MCF-7 cells at 90% confluency using Lipofectamine 2000 transfection reagent in accordance with the protocol of the manufacturer.
MTT assay
The second day after transfection, cells were seeded in 96-well plates with 1,000 cells per well. At the proper time, the cells were incubated with 10 µL 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) (5 mg/mL) for 4 h at 37 °C, subsequently, the MTT medium was removed, and the cells were incubated with 100 µL DMSO for 30 min at room temperature. The absorbance was determined at 490 nm.
Colony formation assay
The MCF-7 cells from different treatments were seeded in 6-well plates, containing 800 cells per well. After 3 weeks of growth, the colonies were washed twice with phosphate-buffered saline (PBS) and fixed in 95% ethanol for 10 min, and then stained with 0.1% crystal violet for 20 min. The number of colonies with more than 50 cells was counted. The experiments were repeated three times in triplicate.
Luciferase reporter assay
The MCF-7 cells were seeded in 24-well plates before transfection. The cells first were transfected with either the miR let-7a mimics or NC mimics. Subsequently, the cells were cotransfected with pMIR-USP32-3'UTRw, pMIR-USP32-3'UTRm or pMIR-REPORT plasmid for 48 h. The cells were harvested and measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
Western blot analysis
Western blotting was performed to detect USP32 protein expression. Whole cell lysates were prepared after transfection using lysis buffer supplemented with protease inhibitors. Equal amounts of extracted protein (40 µg) were separated by a 10% SDS-PAGE. After transferring to polyvinylidene fluoride membrane (Millipore), the membranes were blocked with 5% skim milk for 1 hour at the room temperature. Subsequently, the PVDF membranes were washed 2 times with TBST and incubated overnight at 4 °C with primary antibodies USP32 (1:5,000) and β-actin (1:2,000). Followed by incubation with appropriate secondary antibodies for 2 h at room temperature, and then washed three times with TBST. The bands were detected using BioImaging Systems (UVP Inc., Upland, CA).
Statistical analysis
Data were described as mean ± standard deviation (SD). GraphPad Prism 5.0 software (La Jolla, CA) and SPSS 17.0 software (SPSS Inc. Chicago, IL, USA) were used to analyze the experimental results. Statistical differences between groups were evaluated using Student’s t-test. P<0.05 was considered to be statistically significant.
Results
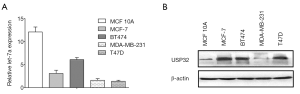
USP32 expression is upregulated in human BCa cells
MiR let-7a and USP32 expression were detected by quantitative reverse transcription polymerase chain reaction (qRT-PCR) and western blot analysis in the selected four human BCa cell lines and one human non-malignant cell line. The qRT-PCR analysis showed that the expression of miR let-7a in non-malignant breast epithelial cells MCF10A was significantly higher than that in four human BCa cell lines (Figure 1A). The Western blot assay revealed that USP32 protein expression was significantly higher in BCa cells than that in non-malignant breast epithelial cells MCF10A (Figure 1B). The above results showed that there is a negatively correlation between the expression of miR let-7a and USP32 protein. Furthermore, the protein expression of USP32 was much higher in MCF-7 BCa cells compared with that in other BCa cells. Therefore, MCF-7 was chosen to explore the relationship between miR let-7a and USP32 expression.
MiR let-7a downregulates USP32 protein expression in MCF-7 cells
To investigate the potential influences of miR let-7a on USP32 protein expression, MCF-7 cells were transfected with miR let-7a mimics, NC mimics, miR let-7a inhibitor or NC inhibitor for 48 h. The total RNA and the whole cell lysates were extracted for qRT-PCR assay and Western blot analysis. The qRT-PCR analysis confirmed the mature let-7a levels in the cells transfected with miR let-7a mimics were significantly higher than that in the cells transfected with NC mimics (Figure 2A).
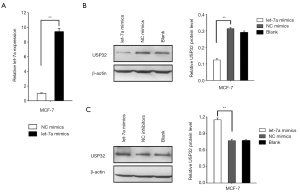
Western blot was performed to detect USP32 protein expression level after transfecting miR let-7a mimics or miR let-7a inhibitors. The results indicated that let-7a mimics could reduce the protein level of USP32 (Figures 2B,S1A), while let-7a inhibitors enhanced the USP32 protein level (Figure 2C). Collectively, the results demonstrated that miR let-7a could downregulate the protein levels of USP32 in MCF-7 cells.
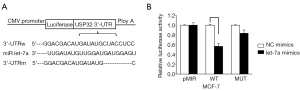
MiR let-7a directly interacts with its targeting sites in the USP32 3'UTR
In order to investigate the molecular mechanisms of miR let-7a acting as a tumor suppressor in BCa, we searched for several potential target genes of let-7a through bioinformatics and selected USP32 as a gene of our interest. The miRNA let-7a targeting sequences were found in the 3'UTR region of USP32 mRNA (Figure 3A) using TargetScan-Human 6.2, and the putative binding site for miRNA let-7a was located at 422-428bp of the USP32-3'UTR. To investigate whether miRNA let-7a can directly bind to this predicted target site in the USP32-3'UTR to downregulate the USP32 protein level, WT 3'UTR (pMIR-USP32-3'UTRw), deletion mutant 3'UTR (pMIR-USP32-3'UTRm) or pMIR-REPORT control plasmids were cotransfected into MCF-7 cells with either t miR let-7a mimics or NC mimics. The reporter assay results (Figure 3B) indicated that overexpression of miR let-7a markedly reduced luciferase activity of the pMIR-USP32-3'UTRw compared with the NC mimics. But had no significant effects on the luciferase activities of the pMIR-USP32-3'UTRm or pMIR-REPORT control plasmids. These data combined with the results in Figure 2 demonstrate that the predicted target site in the USP32 3'UTR was an efficient binding site for miR let-7a, and let-7a reduced USP32 expression through directly targeting the sites in 3'UTR of the USP32 mRNA.
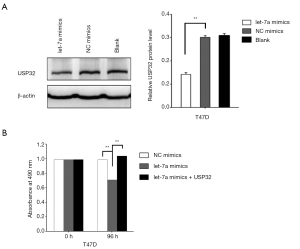
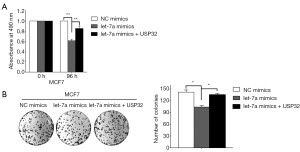
USP32 overexpression alleviates the inhibitory effects of MiR let-7a on cell proliferation
MiR let-7a has been identified as a tumor suppressor to inhibit proliferation in several cancers including BCa. The present study aimed to explore the function of miR let-7a to inhibit MCF-7 cells growth by inhibiting the expression of USP32. After MiR let-7a mimics and NC mimics were transfected into MCF-7 cells for 24 h. USP32 expression plasmid (Myc-USP32) was transfected into MCF-7 cells. Then the cells proliferation of MCF-7 was examined by MTT and colony formation assay. MTT results indicated that the cells transfected with miR let-7a showed significantly reduced cell viability compared with cells transfected with NC mimics, while the cells cotransfected with miR let-7a and USP32 expression plasmid showed a significant increase of cell proliferation compared with the cells transfected with miR let-7a (Figures 4A,S1B). Moreover, colony formation analysis showed that miR let-7a resulted in significantly decrease colonies numbers compared with NC mimics. However, USP32 ectopic expression in miR let-7a group led to a markedly increase colonies numbers in MCF-7 cells (Figure 4B). These results suggest that the effect of miR let-7a on cells proliferation was associated with downregulation of USP32 in the MCF-7 cells.
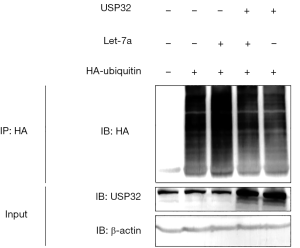
miR let-7a enhances cellular ubiquitination by regulating USP32
Our results have shown that miR let-7a could inhibit BCa cells growth through negatively regulating the expression of USP32. Since USP32 is a member of DUB family and play a pivotal role in regulating protein deubiquitination, thus we wondered whether miR let-7a could affect cellular levels of ubiquitination by targeting USP32. To investigate this hypothesis, the ubiquitination of the whole cellular lysate was detected using an in vivo ubiquitination assay. HA-tagged ubiquitin and miR let-7a were cotransfected into MCF-7 cells in the presence of USP32 expression plasmid or control vector. Immunoprecipitation (IP) with anti-HA antibody was followed by HA immunoblotting. The results showed that overexpression of miR let-7a dramatically increase the cellular ubiquitination, in contrast, overexpression of USP32 markedly reduced the ubiquitination of whole proteins. Overexpression of USP32 alleviated miR let-7a induced enhancement of cellular ubiquitination (Figure 5). Taken together, the results suggest that miR let-7a could increase cellular ubiquitination by decreasing the expression of USP32.
Discussion
Numerous evidences have showed that miRNAs are vital as activators or inhibitors of the developmental process of tumor (22,23). They may exert their function by regulating oncogenes or tumor suppressor genes to modulate tumorigenesis and multiple cellular biological processes of cancer. For example, miR-3191 promoted colorectal cancer cell migration and invasion by downregulating TGFBR2 (24). MiR-552 promoted migration, invasion and MMPs expression of osteosarcoma by targeting TIMP2 (25). Therefore, identification of the target genes is important for exploring the potential mechanisms of miRNAs function in tumor development and progression.
MiR let-7a, as an important tumor suppressor gene, has been demonstrated to suppress tumor growth in gastric cancer, BCa, non-small cell lung cancer and ovarian cancer (26-29). However, previous studies showed the mechanisms by which miR let-7a inhibits cell proliferation is diverse. For example, miR let-7a suppress T-cell growth and IFN-γ secretion by reducing STAT3 protein expression in psoriasis (30). MiR let-7a functions as a tumor suppressor to inhibit cell proliferation, migration and invasion by targeting MAGE-A1 in BCa (31). Overexpression of miR let-7a significantly suppressed osteosarcoma cells proliferation via reducing E2F2 protein expression (32). To further gain insight into the mechanisms underlying miR let-7a function in BCa, we performed a bioinformatics analysis of the target genes of miR let-7a and selected USP32 as a gene of interest.
USP32 localizes on chromosome 17q23 and is a member of the ubiquitin hydrolase family. As a highly conserved gene, USP32 was significantly upregulated in BCa cell lines and breast tissue, and silencing of USP32 causes lower proliferation and migration abilities compared to control (33), and in estrogen receptor-positive tumours, DNA copy number and gene expression of USP32 were increased (34). Moreover, USP32 was found to be overexpressed in human small cell lung cancer tissues, and USP32 silencing significantly suppressed small cell lung cancer cells proliferation and migration, cell cycle progression arrested and cell apoptosis (35). The biological effects and mechanisms underlying USP32 function in tumors remains largely unknown. There has been no data showing the correlations between the expression of USP32 and miRNA.
To validate whether USP32 protein expression is downregulated by miR let-7a, we examined the level of USP32 protein after transfecting miR let-7a mimics and inhibitors. The results revealed that the USP32 protein level was significantly decreased in the miR let-7a mimics group and was greatly increased in miR let-7a inhibitors group. To further verify a direct interaction between miR let-7a and USP32, luciferase reporter gene assay was performed and the result showed that the luciferase activity of pMIR- USP32-3'UTRw but not pMIR-USP32-3'UTRm and pMIR-REPORT plasmids, was markedly suppressed by miR let-7a transfection. Since USP32 functions as a DUB, miR let-7a might affect cellular ubiquitination by targeting USP32. Our data suggest that overexpression of miR let-7a dramatically increase the cellular ubiquitination, and USP32 alleviated miR let-7a induced enhancement of cellular ubiquitination. Taken together, these results confirmed that miR let-7a can inhibit USP32 protein expression level by directly binding USP32 3'UTR.
Our present results showed that USP32 expression is up-regulated in human BCa cells and miR let-7a significantly inhibits USP32 expression. Overexpression of USP32 alleviated miR let-7a induced MCF-7 cells growth inhibition. MiR let-7a enhances cellular ubiquitination by regulating USP32 activity. In conclusion, the present study demonstrated that miR let-7a exhibit a pivotal role in inhibiting proliferation at least partly by regulating the USP32 in BCa, which represents a novel potential therapeutic target for BCa.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.08.30). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional ethical approval and individual informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Das SS, Karmakar P, Nandi AK, et al. Small RNA mediated regulation of seed germination. Front Plant Sci 2015;6:828. [Crossref] [PubMed]
- Mulrane L, McGee SF, Gallagher WM, et al. miRNA dysregulation in breast cancer. Cancer Res 2013;73:6554-62. [Crossref] [PubMed]
- Yanokura M, Banno K, Iida M, et al. MicroRNAS in endometrial cancer: recent advances and potential clinical applications. Excli j 2015;14:190-8. [PubMed]
- Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 2003;113:673-6. [Crossref] [PubMed]
- Kanellopoulou C, Monticelli S. A role for microRNAs in the development of the immune system and in the pathogenesis of cancer. Semin Cancer Biol 2008;18:79-88. [Crossref] [PubMed]
- Schmittgen TD. Regulation of microRNA processing in development, differentiation and cancer. J Cell Mol Med 2008;12:1811-9. [Crossref] [PubMed]
- Wang YY, Ren T, Cai YY, et al. MicroRNA let-7a inhibits the proliferation and invasion of nonsmall cell lung cancer cell line 95D by regulating K-Ras and HMGA2 gene expression. Cancer Biother Radiopharm 2013;28:131-7. [Crossref] [PubMed]
- Tang R, Yang C, Ma X, et al. MiR-let-7a inhibits cell proliferation, migration, and invasion by down-regulating PKM2 in gastric cancer. Oncotarget 2016;7:5972-84. [PubMed]
- Cai K, Wan Y, Sun G, et al. Let-7a inhibits proliferation and induces apoptosis by targeting EZH2 in nasopharyngeal carcinoma cells. Oncol Rep 2012;28:2101-6. [Crossref] [PubMed]
- Liu Y, Yin B, Zhang C, et al. Hsa-let-7a functions as a tumor suppressor in renal cell carcinoma cell lines by targeting c-myc. Biochem Biophys Res Commun 2012;417:371-5. [Crossref] [PubMed]
- Ye Y, Blaser G, Horrocks MH, et al. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature 2012;492:266-70. [Crossref] [PubMed]
- Xiao N, Li H, Luo J, et al. Ubiquitin-specific protease 4 (USP4) targets TRAF2 and TRAF6 for deubiquitination and inhibits TNFalpha-induced cancer cell migration. Biochem J 2012;441:979-86. [Crossref] [PubMed]
- Fraile JM, Quesada V, Rodriguez D, et al. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene 2012;31:2373-88. [Crossref] [PubMed]
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 2009;10:550-63. [Crossref] [PubMed]
- Panier S, Durocher D. Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat Rev Mol Cell Biol 2013;14:661-72. [Crossref] [PubMed]
- Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature 2009;458:430-7. [Crossref] [PubMed]
- Lindner HA. Deubiquitination in virus infection. Virology 2007;362:245-56. [Crossref] [PubMed]
- Sun X, Ding Y, Zhan M, et al. Usp7 regulates Hippo pathway through deubiquitinating the transcriptional coactivator Yorkie. Nat Commun 2019;10:411. [Crossref] [PubMed]
- Lu C, Ning Z, Wang A, et al. USP10 suppresses tumor progression by inhibiting mTOR activation in hepatocellular carcinoma. Cancer Lett 2018;436:139-48. [Crossref] [PubMed]
- Zeng Z, Li D, Yu T, et al. Association and clinical implication of the USP10 and MSH2 proteins in non-small cell lung cancer. Oncol Lett 2019;17:1128-38. [PubMed]
- Palazón-Riquelme P, Worboys JD, Green J, et al. USP7 and USP47 deubiquitinases regulate NLRP3 inflammasome activation. EMBO Rep 2018; [Crossref] [PubMed]
- Lozada-Delgado EL, Grafals-Ruiz N, Miranda-Román MA, et al. Targeting MicroRNA-143 Leads to Inhibition of Glioblastoma Tumor Progression. Cancers (Basel) 2018; [Crossref] [PubMed]
- Wang Y, Wang B, Xiao S, et al. miR-125a/b inhibits tumor-associated macrophages mediated in cancer stem cells of hepatocellular carcinoma by targeting CD90. J Cell Biochem 2019;120:3046-55. [Crossref] [PubMed]
- He H, Zhao X, Zhu Z, et al. MicroRNA-3191 promotes migration and invasion by downregulating TGFBR2 in colorectal cancer. J Biochem Mol Toxicol 2019;e22308. [Epub ahead of print]. [Crossref] [PubMed]
- Chao Y, Hu K, Wang X, et al. MicroRNA-552 promotes migration and invasion of osteosarcoma through targeting TIMP2. Biochem Biophys Res Commun 2019;511:63-8. [Crossref] [PubMed]
- Shekari N, Asghari F, Haghnavaz N, et al. Let-7a Could Serve as A Biomarker for Chemo-Responsiveness to Docetaxel in Gastric Cancer. Anticancer Agents Med Chem 2019;19:304-9. [Crossref] [PubMed]
- Yao A, Xiang Y, Si YR, et al. PKM2 promotes glucose metabolism through a let-7a-5p/Stat3/hnRNP-A1 regulatory feedback loop in breast cancer cells. J Cell Biochem 2019;120:6542-54. [Crossref] [PubMed]
- Qi L, Liu F, Zhang F, et al. lncRNA NEAT1 competes against let-7a to contribute to non-small cell lung cancer proliferation and metastasis. Biomed Pharmacother 2018;103:1507-15. [Crossref] [PubMed]
- Lu L, Schwartz P, Scarampi L, et al. MicroRNA let-7a: a potential marker for selection of paclitaxel in ovarian cancer management. Gynecol Oncol 2011;122:366-71. [Crossref] [PubMed]
- Hu XP, Xie Q, Chen CF, et al. Let-7a Inhibits T-Cell Proliferation and IFN-gamma Secretion by Down-Regulating STAT3 Expression in Patients with Psoriasis. Cell Physiol Biochem 2017;42:115-25. [Crossref] [PubMed]
- Mi Y, Liu F, Liang X, et al. Tumor suppressor let-7a inhibits breast cancer cell proliferation, migration and invasion by targeting MAGE-A1. Neoplasma 2019;66:54-62. [Crossref] [PubMed]
- Iwasaki T, Tanaka K, Kawano M, et al. Tumor-suppressive microRNA-let-7a inhibits cell proliferation via targeting of E2F2 in osteosarcoma cells. Int J Oncol 2015;46:1543-50. [Crossref] [PubMed]
- Akhavantabasi S, Akman HB, Sapmaz A, et al. USP32 is an active, membrane-bound ubiquitin protease overexpressed in breast cancers. Mamm Genome 2010;21:388-97. [Crossref] [PubMed]
- Zhang Y, Martens JW, Yu JX, et al. Copy number alterations that predict metastatic capability of human breast cancer. Cancer Res 2009;69:3795-801. [Crossref] [PubMed]
- Hu W, Wei H, Li K, et al. Downregulation of USP32 inhibits cell proliferation, migration and invasion in human small cell lung cancer. Cell Prolif 2017; [Crossref] [PubMed]


