
Knockdown of actin-like 8 inhibits cell proliferation by regulating FOXM1, STMN1, PLK1, and BIRC5 in lung adenocarcinoma A549 cells
Introduction
Non-small cell lung cancer (NSCLC) is one of the most prevalent malignant diseases in the world. NSCLC includes lung adenocarcinoma (LUAD), lung squamous cell carcinoma, and large cell carcinoma. Among these, LUAD has become the most common subtype of NSCLC (1). With the development of tumor biology in recent years, many tumor target antigens that are involved in the growth, progression, and spread of LUAD have been identified, and include EGFR, ALK, KRAS, etc. (2).
Cancer/testis antigens (CTAs) belong to a class of tumor-related antigens characterized by their spontaneous immunogenicity and the highly tissue-restricted expression (3). They are present in the testicles and various tumor tissues, while having a low expression in other normal adult tissues (3). To date, more than 275 CTA genes have been identified (http://www.cta.lncc.br). Due to their limited expression patterns, CTAs represent a promising candidate for cancer immunotherapy. In recent years, some CTAs like MAGE-A3 and NY-ESO-1 have been studied as target antigens in clinical trials for many cancers, including glioblastoma, NSCLC, melanoma, and ovarian cancer (4-7). Actin-like protein 8 (ACTL8), a member of the CTA family, is highly expressed in glioblastoma, colon adenocarcinoma, breast cancer, endometrial carcinoma, and head and neck squamous cell carcinoma (8-10). Although it is expressed at a low level in the normal bladder, pancreas, thymus, and colon tissues (8), to date, the role of ACTL8 in LUAD remains unclear.
Therefore, we investigated the expression and the role of ACTL8 in LUAD. First, the high expression of ACTL8 was observed in patients with LUAD by immunohistochemistry (IHC) staining. Second, cell proliferation was inhibited by knockdown of ACTL8 in LUAD A549 cells. Third, we performed a global gene expression profile to investigate the potential genes and signal pathways modulated by ACTL8 in A549 cells. Ingenuity pathway analysis (IPA) was administered to display an overview of ACTL8’s possible biological interaction through knockdown of ACTL8 in A549 cells. Lastly, RT-PCR and a Western blot analysis were used to confirm that the expression levels of FOXM1, STMN1, PLK1, and BIRC5 were markedly decreased in ACTL8 knockdown A549 cells.
Methods
Patients and IHC
The tumor tissues and pericarcinomatous (PC) tissues from 45 patients with LUAD were obtained from the China-Japan Friendship Hospital. In addition, 10 frozen normal lung samples were obtained from the tissue bank in the China-Japan Friendship Hospital. The patients had a clear pathological diagnosis according to the American Joint Committee on Cancer staging system. The paraffin-embedded sections of tumor tissues, PC tissues, and normal tissues were analyzed by IHC analysis with ACTL8 antibody (1:500, ab96756, Abcam) following methods previously described (11). IHC scores were calculated and used to determine the expression level of ACTL8.
Design of shRNA for ACTL8 and construction of lentivirus
The shRNA for ACTL8 (shACTL8) and scramble (shCtrl) were purchased from Genechem (Shanghai Genechem Co., Ltd., Shanghai, China). One short-hairpin RNA (shRNA) sequence against a targeted sequence of human ACTL8 (TGGAGATCCTGTTTGAGTT) was screened and transfected into 293T cell lines to generate shACTL8, while shCtrl was used as the negative control. The sequences of shACTL8 and shCtrl were GCTGGAGATCCTGTTTGAGTT and TTCTCCGAACGTGTCACGT, respectively.
Cell culture and infection
Human LUAD A549 cell line was purchased from the American Type Culture Collection (Manassas, VA, USA) and was cultured in RPMI-1640 medium containing 10% fetal bovine serum and 1% penicillin-streptomycin in a 5% CO2 atmosphere at 37 °C. After 72 h of being infected with the lentivirus of shACTL8 and shCtrl, cells were subjected to cellular proliferation analysis and gene expression analysis.
Cellular proliferation analysis
After infection, each experimental group cells were digested with trypsin. The suspended cells were seeded into 96-well dishes. The GFP positive cells were detected by Celigo Image Cytometer daily from day 1 to day 5 after planting cells.
Gene expression analysis
Three biological replicates of H549 cells transduced with shACTL8 or shCtrl were microarrayed. Seventy-two hours after infection, total RNA was extracted with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The RNA quality was assessed using a NanoDrop 2000 spectrophotometer (NanoDrop; Thermo Fisher Scientific, Inc., Wilmington, DE, USA) and Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., USA). Between 50–500 ng of RNA was used to generate amplified RNA (aRNA) using a GeneChip™ 3’IVT Express Kit (Affymetrix, USA) following the manufacturer's protocol. The data of gene expression were detected by GeneChip™ PrimeView™ Human Gene Expression Array (Affymetrix, USA) and subsequent analysis was performed using the Affymetrix Expression Console (EC, version 1.1). Differentially expressed genes (DEGs) were identified with fold changes >2 and FDR <0.05. Protein network analysis of DEGs was performed using Qiagen’s IPA (Qiagen, Redwood City, CA, USA) software, as previously described (12).
Western blot
Cell samples were lyzed by using RIPA buffer (Beyotime, Shanghai, China). Denatured samples (20 µg per lane) were separated by using 10% SDS-PAGE and then transferred to a polyvinylidene difluoride membrane (EMD Millipore, USA). The membranes were incubated with primary antibodies against ACTL8 (1:500, ab96756, Abcam), BIRC5 (1:500, ab469, Abcam), FOXM1 (1:500, ab180710, Abcam), CDK1 (1:1,000, ab32094, Abcam), STMN1 (1:1,000, #9562, CST), CCNB1 (1:1,000, #4138, CST), PLK1 (1:1,000, #4513, CST), STMN1 (1:1,000, #9562, CST) and GAPDH (1:4,000, sc-32233, Santa-Cruz). After washing, secondary antibodies that conjugated to HRP were added and incubated for color development. Densitometry analyses were performed using Quantity One software (Bio-Rad Laboratories, Mississauga, Canada).
qRT-PCR
One µg of RNA was reversed transcribed into cDNAs using the M-MLV kit (Promega, Madison, WI, USA). Then, qRT-PCR analysis was performed on the LightCycler 480 system, with the primers listed in Table S1 and the LightCycler 480 SYBR Green Master kit (Roche, Mannheim, Germany) according to the manufacturer’s instructions. GAPDH was used as the endogenous control gene. The relative mRNA abundance analyses were performed using the 2-ΔΔCt analysis method (13). The thermocycling conditions were as follows: 30 sec at 95 °C, followed by 40 cycles of 5 sec at 95 °C, and 30 sec at 60 °C. The last step was conducted at 95 °C for 15 s, 60 °C for 30 s, and 95 °C for 15 s, which is necessary to acquire a melting curve for the PCR products. All assays were performed in triplicate.
Table S1
| Gene | Forward | Reverse | Size (bp) |
|---|---|---|---|
| ACTL8 | GAACATCGTGAACTACCTACCG | CAAGGGTGTCTCCGTGATGAT | 226 |
| GAPDH | TGACTTCAACAGCGACACCCA | CACCCTGTTGCTGTAGCCAAA | 121 |
| AURKB | CAGAAGAGCTGCACATTTGACG | CCTTGAGCCCTAAGAGCAGATTT | 136 |
| TPX2 | TCCTGCCCGAGTGACTAAGG | CTGTTAGGGGTTCGTTTATGGAA | 144 |
| CDK1 | GGATGTGCTTATGCAGGATTCC | CATGTACTGACCAGGAGGGATAG | 100 |
| DHFR | CTGTCTTAGATTGGGGAACCC | ATGAGAACCTGCTCGCTGA | 158 |
| SQSTM1 | GACTACGACTTGTGTAGCGTC | AGTGTCCGTGTTTCACCTTCC | 139 |
| CCNE2 | GGAACCACAGATGAGGTCCAT | CCATCAGTGACGTAAGCAAACT | 237 |
| TRIP13 | ACTGTTGCACTTCACATTTTCCA | TCGAGGAGATGGGATTTGACT | 173 |
| E2F8 | AAGTACGCCGAGCAGATTATG | ATGTCTGGGTGTCCATTTGGG | 128 |
| PRC1 | ATTCATGGAGTATGTGGCAGAAC | AGCATCTCTGTCTCTGTCTGTT | 108 |
| RAD51 | AGCGATGTTTGCTGCTGATCC | TGGTTTCCCCTCTTCCTTTCC | 101 |
| CDKN1A | CTGTCACTGTCTTGTACCCTTGT | AAATCTGTCATGCTGGTCTGC | 113 |
| FOXM1 | TGGAGCAGCGACAGGTTAAG | AAGGTTGTGGCGGATGGAGT | 233 |
| CENPF | TCGTTCCATCCCTGTCATC | TCCTGGTCAGATTCTCCTCC | 201 |
| E2F7 | TCAGATTCCACAGACCTTGCC | GCTCTTTGGTGCTTTCATTTTC | 211 |
| POLA1 | AGCCAGTCAGTTGGTGTAAAGT | GCTGCCATAGCAATAATCTCAT | 160 |
| UBE2C | GATGTCTGGCGATAAAGGG | TGATAGCAGGGCGTGAGGA | 183 |
| MCM7 | TAACTGTGCGTGGAATCGT | GAGGCATGAAAGTGGGAGA | 126 |
| NEK2 | CAGCCCTGTATTGAGTGAGC | GAACTTCCGTTCCTTTAGCA | 193 |
| RACGAP1 | TCCTTGCCTCTGGAGTATTG | AAGTTGCCTTGTCGTCCTA | 278 |
| CDK2 | CTGGACACTGAGACTGAGG | GAGGACCCGATGAGAATGG | 264 |
| STMN1 | TGAGAAACGAGAGCACGAG | TCAGCAGGGTCTTTGGATTC | 213 |
| CCNB2 | CAACCCACCAAAACAACA | AGAGCAAGGCATCAGAAA | 163 |
| CDC20 | CTTCGGCTCAGTGGAAAA | GTCTGGCAGGGAAGGAAT | 129 |
| MKI67 | GGAACAGCCTCAACCATCAG | CCACTCTTTCTCCCTCCTCTC | 210 |
| TFDP1 | CCCCAGCACTCACTTTG | TCGTCTGCCACTTCGTT | 174 |
| CDCA3 | ATTGCACGGACACCTATGA | TGTGGGCTGTCTTGCTTC | 261 |
| BUB1 | AACCCACAGGAGCCAGGAC | GTGGAATGGTGTAGACGCAAG | 200 |
| BIRC5 | CAAGGACCACCGCATCTC | CCAAGGGTTAATTCTTCAAACT | 255 |
| CCNB1 | CTAAGATTGGAGAGGTTGATGTC | GGTAATGTTGTAGAGTTGGTGTC | 177 |
| PLK1 | AGGCAAGAGGAGGCTGAG | GGATGAGGCGTGTTGAGTC | 145 |
Statistical analysis
Statistical analysis was carried out by the SPSS 19.0 statistics software (SPSS, Inc., Chicago, IL, USA). Data are given as mean ± standard deviation (SD). A student’s t-test (two-sided, for two group samples) and one-way ANOVA (for multiple group samples) were used to analyze the differences among samples. P values <0.05 were considered to be statistically significant.
Results
High expression of ACTL8 in LUAD patients
To detect the expression of ACTL8 in LUAD, we collected 45 tumor tissues with paired PC tissues, and 10 normal lung tissues. The IHC data revealed that the expression of ACTL8 was significantly increased within tumor tissues compared with the PC tissues and normal lung tissues (Figure 1A,B). Of the 45 patients, 30 were less than 56 years of age, and 15 were more than 56 years of age. The male-to-female ratio was 5:4 (25 males: 20 females). In regards to the clinical-stage, 34 were in stage I/II, and 11 were in stage III/IV. The pathology grade was known for 40 cases; 29 were in grade 1/2, and 11 were in grade 3. No significant differences were observed between the expression of ACTL8 and age, gender, clinical stage, pathology grade, and TNM stage (Table 1).
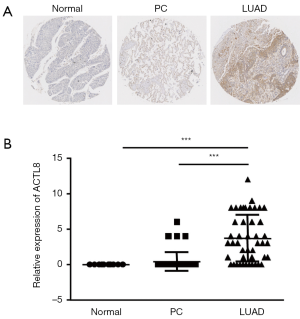
Table 1
| Characteristics | Number | ACTL8 expression | P | |
|---|---|---|---|---|
| Low | High | |||
| Age | ||||
| ≤56 | 30 | 24 | 6 | 0.332 |
| >56 | 15 | 10 | 5 | |
| Gender | ||||
| Male | 25 | 18 | 7 | 0.611 |
| Female | 20 | 16 | 4 | |
| Clinical stage | ||||
| I/II | 34 | 26 | 8 | 0.840 |
| III/IV | 11 | 8 | 3 | |
| Pathology grade | ||||
| N/A | 5 | 4 | 1 | |
| 1/2 | 29 | 22 | 7 | 0.840 |
| 3 | 11 | 8 | 3 | |
| TNM stage | ||||
| TNM1/2 | 38 | 29 | 9 | 0.785 |
| TNM3/4 | 7 | 5 | 2 | |
LUAD, lung adenocarcinoma; ACTL8, actin-like protein 8.
Knockdown of ACTL8 inhibited cell proliferation
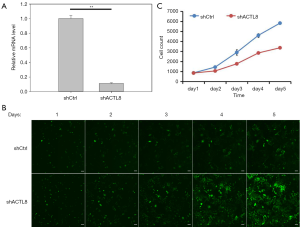
Given the oncogenic role of ACTL8 in breast cancer, we sought to determine whether ACTL8 proteins regulate cell proliferation in LUAD. A549 cells were transfected with the lentivirus of shACTL8 or shCtrl and evaluated by fluorescence microscopy. After 72 h of infection, the level of ACTL8 mRNA expression in cells infected with the shACTL8 was significantly decreased (88.6%) compared with those infected with the shCtrl (P<0.01; Figure 2A). These results show that the knockdown of ACTL8 was effective in A549 cells. Celigo Image Cytometer determined the proliferation ability of A549 cells infected with the shACTL8 or shCtrl. As shown in Figure 2B,C, obviously inhibited proliferation was observed in the shACTL8 group as compared to the shCtrl group.
Gene expression profiles and IPA
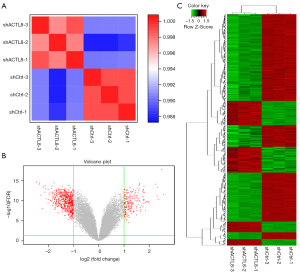
We further performed a microarray to determine the potential genes and pathways regulated by ACTL8 in A549 cells. Pearson’s correlation coefficient between the biological duplicates was >0.99, showing the efficiency of the microarray and confirming the relevance of the samples (Figure 3A). A total of 504 DEGs (146 up-regulated, and 358 down-regulated) were found in the ACTL8 knockdown A549 cells compared with the mock-transfected cells (Figure 3B and http://fp.amegroups.cn/cms/tcr.2019.09.33-1.xlsx). All DEGs were clustered into 8 main groups according to their expression pattern similarity (Figure 3C). All samples were clustered into 2 groups corresponding to shACTL8 and shCtrl, which was consistent with that of the Pearson’s correlation analysis.
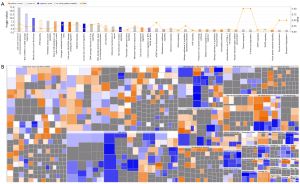
To understand the functions of these DEGs, IPAs were performed and revealed that several canonical pathways such as cyclins and cell cycle regulation, estrogen-mediated S-phase entry, and aryl hydrocarbon receptor signaling, were significantly inhibited, while pathways such as cell cycle G2/M DNA damage checkpoint regulation and HMGB1 signaling, were significantly activated by ACTL8 knockdown (Figure 4A and http://fp.amegroups.cn/cms/tcr.2019.09.33-2.xlsx). Also, the disease and function enrichment analysis revealed that a total of 63 biological functions were significantly altered, with 35 significantly inhibited and 28 activated (Figure 4B and http://fp.amegroups.cn/cms/tcr.2019.09.33-3.xlsx). Of note, many biological functions involved in tumors like cell proliferation of tumor cell lines, metabolism of DNA, DNA replication, M phase of tumor cell lines, and growth of tumor, were significantly inhibited. Meanwwhile, processes associated with cell death and apoptosis were significantly activated (Figure 4B and http://fp.amegroups.cn/cms/tcr.2019.09.33-3.xlsx).
In addition, upstream modulators were predicted by IPA. The results showed that 198 regulators were significantly changed, with 64 being inhibited and 134 activated (http://fp.amegroups.cn/cms/tcr.2019.09.33-4.xlsx). Among these, NUPR1 was the most activated, with 76 genes being uniformly affected. Meanwhile CSF2 was the most inhibited, with 58 genes being uniformly affected (http://fp.amegroups.cn/cms/tcr.2019.09.33-4.xlsx).
Based on the DEGs regulated by CSF2 and DEGs related to cyclins and cell cycle regulation, estrogen-mediated S-phase entry, and aryl hydrocarbon receptor signaling, the interaction network of these DEGs was integrated (Figure 5). The results showed that the expression of CCNB1, PLK1, FOXM1, CDK1, etc. was inhibited by ACTL8 knockdown in A549 cells.
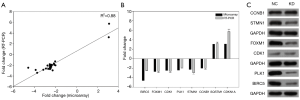
Validation of gene expression array by qRT-PCR and Western Blot
Based on the interaction network analysis, 25 DEGs were selected to perform to qRT-PCR. We calculated the correlation efficiency of the microarray and qRT-PCR, and observed a strong correlation between the two methods (R2=0.88) (Figure 6A). The qRT-PCR results also showed that the mRNA expression of CDKN1A and SQSTM1 were up-regulated following ACTL8 knockdown, whereas CCNB1, CDK1, STMN1, FOXM1, PLK1, and BIRC5 were down-regulated (Figure 6B). Also, the protein expression of CCNB1, CDK1, STMN1, FOXM1, PLK1, and BIRC5 were detected by Western blotting. The results demonstrated that knockdown of ACTL8 decreased the level of FOXM1, STMN1, PLK1, and BIRC5 (Figure 6C).
Discussion
With the development of tumor biology in recent years, specific targeted therapy and immunotherapy have been available for the treatment of advanced LUAD (2). CTA family members are becoming increasingly promising as targets for cancer immunotherapy approaches (14-16). As a member of the CTA family, the expression of ACTL8 has significantly increased in many cancers. However, the role of ACTL8 in LUAD remains unknown.
In this study, we firstly found that ACTL8 was highly expressed in human LUAD tissues. Secondly, a shACTL8 lentiviral vector was constructed. Significant decrease in proliferation was observed in the shACTL8 group through RNA interference. We then performed microarray analysis and IPA to investigate the potential genes and signal pathways modulated by ACTL8 in A549 cells. The results reveal a set of 504 DEGs, a number of which can be categorized into cyclins and cell cycle regulation and estrogen-mediated S-phase functions, which were significantly inhibited. In contrast, cell cycle G2/M DNA damage checkpoint regulation was the most activated pathway. These results provide a plausible explanation for the biological phenomena as described above. In addition, for disease and function analysis, processes associated with cell proliferation of tumor cell lines, metabolism of DNA, and DNA replication, were significantly inhibited. However, cell death and apoptosis were significantly activated.
Upstream regulator analysis by IPA showed that NUPR1 was the most activated. NUPR1 is a transcription factor which plays a crucial role in the cell cycle and cell death (17). Many studies have shown that NUPR1 plays a vital role in cancer development and progression (17). In NSCLC, knockdown of NUPR1 inhibits cell proliferation and colony formation in vitro and in vivo (18).
Finally, 25 candidate genes were selected for validation of gene expression using RT-qPCR. Among these, 6 genes, including CCNB1, CDK1, STMN1, FOXM1, PLK1, and BIRC5, were further detected by Western blot. The results showed that the protein expression levels of FOXM1, STMN1, PLK1, and BIRC5 were markedly reduced in ACTL8 knockdown A549 cells, which was consistent with the microarray analysis. FOXM1 is a transcriptional factor regulating the expression of cell cycle genes essential for DNA damage repair, cell proliferation, cell differentiation, and tissue homeostasis (19). It plays an important role in G2/M progression in normal human cells (20). Its transcriptional targets include CCNB1 (21), BIRC5 (22), PLK1 (23), and STMN1 (24). FOXM1 transcriptional activity also requires phosphorylation by CDK1 and PLK1 (23). PLK1 is an erine/threonine-protein kinase that performs several important functions throughout the M phase of the cell cycle. STMN1 is a microtubule-destabilizing phosphoprotein involved in the tumor cell cycle (25) and metastasis (26). A recent study reveals that activation of an AKT/FOXM1/STMN1 pathway drives resistance to tyrosine kinase inhibitors in lung cancer (24).
Conclusions
Taken together, the results of the present study reveal that ACTL8 is highly expressed in LUAD tissues and promotes cell proliferation by regulating FOXM1, STMN1, PLK1, and BIRC5 in A549 cells. However, the exact underlying molecular mechanisms are yet to be determined.
Acknowledgments
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.33). The authors have no conflicts of interest to declare
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent from all patients and the approval from the Medical Ethics Committee of China-Japan Friendship Hospital have been obtained (the ID number is 170205).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Rothschild SI. Targeted Therapies in Non-Small Cell Lung Cancer-Beyond EGFR and ALK. Cancers (Basel) 2015;7:930-49. [Crossref] [PubMed]
- Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: review, standardization, and commentary. Cancer Immun 2004;4:1. [PubMed]
- Phuphanich S, Wheeler CJ, Rudnick JD, et al. Phase I trial of a multi-epitope-pulsed dendritic cell vaccine for patients with newly diagnosed glioblastoma. Cancer Immunol Immunother 2013;62:125-35. [Crossref] [PubMed]
- Odunsi K, Matsuzaki J, Karbach J, et al. Efficacy of vaccination with recombinant vaccinia and fowlpox vectors expressing NY-ESO-1 antigen in ovarian cancer and melanoma patients. Proc Natl Acad Sci U S A 2012;109:5797-802. [Crossref] [PubMed]
- Vansteenkiste J, Zielinski M, Linder A, et al. Adjuvant MAGE-A3 immunotherapy in resected non-small-cell lung cancer: phase II randomized study results. J Clin Oncol 2013;31:2396-403. [Crossref] [PubMed]
- Robbins PF, Morgan RA, Feldman SA, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol 2011;29:917-24. [Crossref] [PubMed]
- Freitas M, Malheiros S, Stávale JN, et al. Expression of cancer/testis antigens is correlated with improved survival in glioblastoma. Oncotarget 2013;4:636-46. [Crossref] [PubMed]
- Yao J, Caballero OL, Yung WKA, et al. Tumor subtype-specific cancer-testis antigens as potential biomarkers and immunotherapeutic targets for cancers. Cancer Immunol Res 2014;2:371-9. [Crossref] [PubMed]
- Li B, Zhu J, Meng L. High expression of ACTL8 is poor prognosis and accelerates cell progression in head and neck squamous cell carcinoma. Mol Med Rep 2019;19:877-84. [PubMed]
- Liu L, Li D, Chen S, et al. B7-H4 expression in human infiltrating ductal carcinomaassociated macrophages. Mol Med Rep 2016;14:2135-42. [Crossref] [PubMed]
- Krämer A, Green J, Pollard J Jr, et al. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30:523-30. [Crossref] [PubMed]
- Kubista M, Andrade JM, Bengtsson M, et al. The real-time polymerase chain reaction. Mol Aspects Med 2006;27:95-125. [Crossref] [PubMed]
- Dhodapkar MV, Osman K, Teruya-Feldstein J, et al. Expression of cancer/testis (CT) antigens MAGE-A1, MAGE-A3, MAGE-A4, CT-7, and NY-ESO-1 in malignant gammopathies is heterogeneous and correlates with site, stage and risk status of disease. Cancer Immun 2003;3:9. [PubMed]
- Caballero OL, Chen YT. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci 2009;100:2014-21. [Crossref] [PubMed]
- Garcia-Soto AE, Schreiber T, Strbo N, et al. Cancer-testis antigen expression is shared between epithelial ovarian cancer tumors. Gynecol Oncol 2017;145:413-9. [Crossref] [PubMed]
- Cano CE, Hamidi T, Sandi MJ, et al. Nupr1: the Swiss-knife of cancer. J Cell Physiol 2011;226:1439-43. [Crossref] [PubMed]
- Guo X, Wang W, Hu J, et al. Lentivirus-mediated RNAi knockdown of NUPR1 inhibits human nonsmall cell lung cancer growth in vitro and in vivo. Anat Rec (Hoboken) 2012;295:2114-21. [Crossref] [PubMed]
- Zona S, Bella L, Burton MJ, et al. FOXM1: an emerging master regulator of DNA damage response and genotoxic agent resistance. Biochim Biophys Acta 2014;1839:1316-22. [Crossref] [PubMed]
- Down CF, Millour J, Lam EW, et al. Binding of FoxM1 to G2/M gene promoters is dependent upon B-Myb. Biochim Biophys Acta 2012;1819:855-62. [Crossref] [PubMed]
- Leung TW, Lin SS, Tsang AC, et al. Over-expression of FoxM1 stimulates cyclin B1 expression. FEBS Lett 2001;507:59-66. [Crossref] [PubMed]
- Wang IC, Chen YJ, Hughes D, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol 2005;25:10875-94. [Crossref] [PubMed]
- Fu Z, Malureanu L, Huang J, et al. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol 2008;10:1076-82. [Crossref] [PubMed]
- Li M, Yang J, Zhou W, et al. Activation of an AKT/FOXM1/STMN1 pathway drives resistance to tyrosine kinase inhibitors in lung cancer. Br J Cancer 2017;117:974-83. [Crossref] [PubMed]
- Rubin CI, Atweh GF. The role of stathmin in the regulation of the cell cycle. J Cell Biochem 2004;93:242-50. [Crossref] [PubMed]
- Wu W, Tan XF, Tan HT, et al. Unbiased proteomic and transcript analyses reveal that stathmin-1 silencing inhibits colorectal cancer metastasis and sensitizes to 5-fluorouracil treatment. Mol Cancer Res 2014;12:1717-28. [Crossref] [PubMed]


