
DNA hypermethylation of the promoter attenuates forkhead box protein 3 (FOXP3) expression in hepatocellular carcinoma cells
Introduction
Forkhead box protein 3 (FOXP3) was first discovered in samples of tissues from scurfy mice with X-linked recessive genetic immune disorders and severe autoimmune diseases. FOXP3 is an important member of the forkhead/winged helix family of transcription factors (1). In previous studies, FOXP3 has had high relative levels of expression in regulatory T cells (Treg), has been indicated as the decisive factor in the development of and for proper function of Treg cells, and was identified as the most characteristic molecular marker indicative of the presence of Treg cells (2,3). Recent studies have indicated that FOXP3 occurs in and effects the growth and development of different types of tumor cells, including of pancreatic cancer (4), colorectal cancer (5,6), malignant melanoma (7,8), thyroid carcinoma (9), liver carcinoma (10), and ovarian carcinoma (11). Epigenetic related modifications have been deemed as potentially important mechanisms which can help regulate reregulate FOXP3 expression and function within Treg cells (12). However, few reports are available which have examined epigenetics as related to FOXP3 expression in tumor cells and methylation studies of FOXP3 in tumor cells have yet to be published. Thus, the main purpose of this study was to examine and discuss the role of FOXP3 gene promoter methylation in hepatocellular carcinoma (HCC) afflicted cells. The new information should help lay an initial foundation and route for further studies of the role of epigenetics and FOXP3 genes in HCC dynamics.
Methods
HCC tissue samples
Eight pairs of HCC afflicted samples and of corresponding samples of nearby non-tumorous tissues were collected between 2016 and 2017 from eight individual HCC patients who underwent primary and curative tumor resectioning in our hospital. All subjects were provided with, and signed written informed consent prior to sample collection. Approvals related to human ethics guidelines for this study were obtained from the Clinical Research Ethics Committee at Shenzhen Hospital, Peking University, China.
Cell lines and cell culture
Non-tumorous liver cells of line LO2 and HCC afflicted cells of cell lines Huh7 and BEL7402 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cell line HepG2 was provided by Dr. Liu Zhigao from the laboratory of Shenzhen PKU-HKUST Medical Center. The SMMC7721 cell line was provided by Dr. Shi Min from Reproductive Medical Laboratory of Shenzhen PKU-HKUST Medical Center. All cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal bovine serum (FBS) in a 5% CO2 atmosphere at 37 °C. We also used 100 U/mL penicillin, and streptomycin as supplements for the medium.
RT-PCT and real-time quantitative PCR
Total RNA was extracted using Trizol reagent (Invitrogen) following manufacturer protocols. Reverse transcription was performed using the PrimeScript RT reagent Kit (Takara, Japan) following all manufacturer protocols. Quantitative RT-PCR was performed using SYBR Prime Script RT-PCR Kits (Takara, Japan) following all manufacturer protocols. Expression levels of FOXP3 were calculated with the 2-ΔΔCt method normalized using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA. All assays were performed in triplicate. The primer sets we used for examination of the levels of expression were as follows: FOXP3 forward primer, 5'-GGCCCTTCTCCAGGACAGA-3'; FOXP3 reverse primer, 5'-GCTGATCATGGCTGGGTTGT-3'; GAPDH forward primer, 5'-AGAAGGCTGGGGCTCATTTG-3'; and GAPDH reverse primer, 5'-AGGGGCCATCCACAGTCTTC-3'.
Immunohistochemistry assay
Hepatocellular tissue samples were fixed in 10% formalin and embedded in paraffin overnight. Tissues were sectioned at 4 mm thickness and were deparaffinized and rehydrated through a xylene and graded alcohol series. Samples of sectioned tissue were then boiled in citric acid buffer. Samples of sections afflicted by HCC were incubated overnight at 4 °C with the anti-FOXP3 antibody. After washing in PBS, samples were incubated at 37 °C with a second peroxidase reagent for 60 min, followed by 3,3'-diaminobenzidine (DAB) substrate staining, and counterstaining with Vector Hematoxylin QS. Resultant images were captured using microscopy (Olympus BX41).
Western blot
For protein extraction, cultured HCC afflicted cell lines were lysed in cell lysis buffer including a protease inhibitor cocktail (Roche). Then, protein samples were separated using 10% SDS PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore Corp., USA). Transfers were followed by blocking with 5% bovine serum albumin (BSA) for 2 hrs at room temperature and incubation with the primary antibody at 4 °C overnight. After washing with TBST 3 times, membranes were incubated with peroxidase conjugated secondary antibodies for 1 hr at room temperature. Amplified band intensities were determined using chemiluminescent reagents (ThermoFisher Scientific, USA).
DNA methylation analysis
We predicted the occurrences of CpG islands using the UCSC Genome Browser. Genomic DNA from LO2 and 7721 cells was prepared using the MethylDetector kit (Active Motif Co.). The primer set for PCR was as follows: forward primer, 5'-TTTATTTGGGTTAAGTTTGTTGTAGG-3'; reverse primer, 5'-CATCTAAACCCTATTATCACAACCC-3'. PCR products were cloned using a T4 ligase cloning kit (Takara), and single clones were sent for sequencing. The levels of expression of FOXP3 in 7721 cells treated with or without 3 mM 5-aza-2'-deoxycytidine (5-AzadC; Sigma) for 72 hrs was analyzed by qRT-PCR.
Results
FOXP3 was down-regulated in HCC tissues
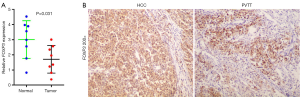
We investigated levels of expression of FOXP3 in HCC afflicted tissues and corresponding adjacent non-tumorous liver tissues sampled from 8 individual HCC patients. Using real-time PCR, we found that the levels of mRNA expression of FOXP3 in HCC tissues were significantly decreased compared with the corresponding levels in adjacent non-tumorous liver tissues (P=0.031; Figure 1A). Immunohistochemistry assays which examined patient levels of FOXP3 protein expression between tumor tissues with (PVTT) and without (HCC) metastasis indicated that levels of expression of FOXP3 proteins in PVTT tissues were significantly lower than were respective levels in HCC afflicted tissues (Figure 1B).
FOXP3 was aberrantly expressed in HCC cell lines
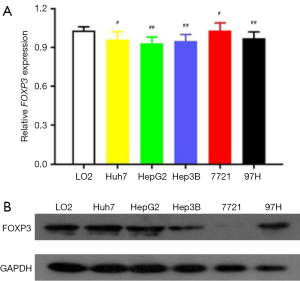
For further confirmation of observed levels of expression of FOXP3 in HCC afflicted samples, we measured the levels of mRNA and proteins of FOXP3 in 5 HCC cell lines (Huh-7, HepG2, Hep3B, 7721, and 97H) and in one non-tumorous human liver cell line (LO2). As demonstrated in Figure 2A, the levels of expression of FOXP3 in HCC afflicted cell lines were lower than respective levels in the non-tumorous liver cell line (Figure 2A). Consistent with the observations for levels of mRNA, results from western blotting revealed a similar decrease in the levels of expression of FOXP3 in HCC afflicted cell lines, compared with the non-tumorous liver cell line LO2 (Figure 2B).
FOXP3 promoter was aberrantly expressed in HCC cell lines
Our results indicated that FOXP3 was abnormally expressed in both HCC afflicted tissues and cell lines, and it has been previously hypothesized that the FOXP3 promoter plays a pivotal role in the regulation of FOXP3 transcription and expression. Therefore, we investigated the role and influence of the FOXP3 promoter region to analyze the corresponding levels of expression of FOXP3 among HCC afflicted cell lines using RT-PCR. As Figure 3 indicates, the FOXP3 promoter was abnormally expressed in HCC afflicted cell lines.

DNA methylation regulates the expression of FOXP3
Previous studies have revealed that DNA hyper-methylation can have a negative effect on the regulation of gene expression. Further, the FOXP3 promoter region was hyper-methylated when multiple types of cancers were analyzed. Thus, we analyzed the methylation status of the FOXP3 promoter by sequencing HCC afflicted cells treated with (methylated cells) or without 5-Aza-CdR (demethylated cells). We collected 10 colonies from methylation-screened LO2 and 7721 cells. Bacterial colonies were propagated using colony PCR and then randomly selected from colonies. Bisulfite sequencing PCR was used to analyze the methylation status of HCC afflicted cells. 7721 cells contained fewer methylated CpG sites than were identified for LO2 cells (Figure 4A). Furthermore, qRT-PCR results indicated that the levels of expression of FOXP3 in 7721 cells were increased approximately 4-fold in the treatment with the addition of 5-Aza-dC (Figure 4B). Such a result suggested that levels of DNA methylation may play an important functional role in the realized levels of expression of FOXP3.

Discussion
FOXP3 is a chromosomal X-linked nuclear transcription factor discovered in efforts resulting from cloning of the Scurfin gene (related to X-linked IPEX autoimmune disease) locus. FOXP3 is also a newly recognized member of the forkhead/winged helix transcription factor family located within chromosome Xp11.23 and includes 11 exons and 10 introns. The total length of the associated cDNA is 1,896 bp (13). Initially, FOXP3 was identified as a surface marker and characterized as a significant a regulatory molecule for CD4+CD25+ Treg cells (14,15). Epigenetics modification is an important consideration in the factors that regulate the levels of FOXP3 expression and has an important function in CD4+CD25+ Treg cells (16,17). Means by which epigenetic modifications can occur include DNA methylation and histone acetylation among others, however, DNA methylation has been the primary focus of study thus far (18). DNA methylation and demethylation often occur on the CpG island region found in DNA promoter sequences (19,20). If the CpG island is methylated chromatin may be condensed, whereas if CpG is demethylated chromatin may be loosened, and such factors are important considerations for possibly increasing accessibility and the ability to target DNA sequences that connect to specific transcription factors (21). Studies have found that in CD4+CD25+ Treg, the CpG sequence motif in the FOXP3 promoter region is nearly classified as demethylated (4,17).
Changes in the levels of DNA methylation are closely related to tumor development and progression. Many methylatation sensitive genes can have abnormal levels of methylation with corresponding abnormal expression in tumorous cells. These are important considerations as related to tumorigenesis, including for tumor suppressor genes, oncogenes, apoptosis-related genes, mismatch repair genes, and others important in cancer related pathways (22). For example, the tumor suppressor gene p16 can be inactivated due to high levels of methylation of the promoter as documented in HCC, ultimately leading to liver cancer (23). Over just the last two years, the relationship between tumor cell dynamics and FOXP3 has attracted increasing attention. FOXP3 is expressed in cells of tumorous cell lines and of tumorous tissues, including for gastric cancer (24), breast cancer (25), lung cancer (26), oral squamous cell carcinoma (27), glioma (28), and prostate cancer (29). Our study demonstrated that the functions and functional effects of FOXP3 are different in different tissues. For example, silencing FOXP3 in melanoma cells by using RNA interference successfully reduced the proliferation of tumorous cells (8). Likewise, when FOXP3 was regulated in a test case with ovarian cancer, cell proliferation was inhibited, cell migration was reduced, and cell invasion was slowed or eliminated (11). Such findings illustrate that FOXP3 could, and likely does affect the development and progression of carcinomas as an independent factor free from synergisms with other factors. However, few studies have thus far been conducted to examine molecular regulatory mechanisms affecting expression and function of FOXP3 in tumorous cells and its synergisms with other mechanisms. Likewise, few studies have examined the role of epigenetic influences upon FOXP3 dynamics in tumor cells, especially as related to DNA methylation inducing tumor onset, development, and progression.
Although few detailed examinations of the roles and influence that FOXP3 plays in the development and progression of tumorous tissues and cells have been undertaken, there has been an increased recognition of the great potential to use FOXP3 as a means for targeted treatment of tumors. According to our findings the Fv region of monoclonal antibodies marked by FOXP3 fusion proteins if targeted could potentially cause dose-dependent cell death for breast cancer, ovarian cancer, and colon cancer. Associated research indicated that antibody-based therapies could reduce neoplasmic hepatic metastases in colon tissues of Balb/c mice, which implies a related cure for cancer could be plausible (30). However, it should be noted that the potentially new therapeutic method of using FOXP3 as a treatment target still requires additional rigorous research and study to fully understand important molecular dynamics and regulatory mechanisms. In our view, although the levels of FOXP3 expression and its functions are quite different from those of Treg cells, epigenetic modifications, and especially methylation related modifications, are still the main approaches with the highest potential to regulate FOXP3 functions and mitigate related effects in tumor cells. In short, an approach considering FOXP3 and the role of epigenetics presents a myriad of broad prospects for further research and application in clinical practice.
In our study, we found that the levels of expression of the FOXP3 gene in liver tissues afflicted by cancer were lower than that for respective levels in adjacent non-tumorous otherwise normal liver tissues. Further, levels of expression of FOXP3 proteins in portal vein tumor thrombus (PVTT) were much lower than respective levels in HCC afflicted samples without metastasis. Consistent with results in sampled tissues, the levels of FOXP3 expression were also partially decreased in HCC afflicted cell lines, compared to the levels observed in the benign unafflicted liver cell line (LO2). We also determined the levels of expression of the FOXP3 promoter during analysis of several HCC afflicted cell lines. The results indicated that the promoter for FOXP3 was abnormally expressed in HCC afflicted cell lines. Further, sequencing was performed to detect the levels of methylation in the FOXP3 gene promoter region of HCC cell line SMMC7721 and LO2 for treatments with (methylated) or without 5-Aza-CdR (demethylated) applied. Results indicated that the frequency of methylation of the FOXP3 promoter region in the SMMC7721 cell line was much higher than were respective levels in the LO2 cell line. This result was confirmed in that the levels of expression of FOXP3 were depressed in SMMC7721 cells, compared to respective levels in LO2 cells. This stood out in contrast to the observation of complete demethylation of CpG sequence motifs in the Treg FOXP3 promoter region (31), and essentially revealed that hypermethylation occurs in the FOXP3 gene promoter region in HCC cells. After reversing the hypermethylation status of the promoter, we were unable to make conclusive statements about the impacts on FOXP3 expression and HCC cell proliferation and invasion and these aspects require further study to reveal important associated dynamics. Thus, our experiment has provided novel information and has helped to lay a foundation for examination of the roles of FOXP3 and epigenetics of HCC cells.
In summary, we found that FOXP3 was down-regulated in HCC afflicted tissues, compared with adjacent non-tumorous tissues. Further, we found that the levels of expression of FOXP3 in PVTT were much lower than respective levels in HCC afflicted tissues. FOXP3 was also abnormally expressed in HCC cell lines. The FOXP3 promoter was also abnormally expressed in the different HCC cell lines. Finally, the FOXP3 promoter was hypermethylated in HCC afflicted cell lines. Our novel results indicate that the levels of expression of FOXP3 mRNA and protein in HCC afflicted tissues and cell lines were decreased. The FOXP3 gene promoter region thus, when hypermethylated, may induce the inhibition of FOXP3 expression and HCC development.
Acknowledgments
Funding: This work was supported by:
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.09). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Tissues were collected from patients. This study was approved by the Medical Ethics Committee of The Shenzhen Hospital, Peking University. Written informed consent was collected from each patient. The current study has been performed in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chatila TA, Blaeser F, Ho N, et al. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest 2000;106:R75-81. [Crossref] [PubMed]
- Sakaguchi S. Regulatory T cells: key controllers of immunologic self-tolerance. Cell 2000;101:455-8. [Crossref] [PubMed]
- Sakaguchi S. Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat Immunol 2005;6:345-52. [Crossref] [PubMed]
- Hinz S, Pagerols-Raluy L, Oberg HH, et al. Foxp3 expression in pancreatic carcinoma cells as a novel mechanism of immune evasion in cancer. Cancer Res 2007;67:8344-50. [Crossref] [PubMed]
- Stanilov N, Miteva L, Mintchev N, et al. High expression of Foxp3, IL-23p19 and survivin mRNA in colorectal carcinoma. Int J Colorectal Dis 2009;24:151-7. [Crossref] [PubMed]
- Kim M, Grimmig T, Grimm M, et al. Expression of Foxp3 in colorectal cancer but not in Treg cells correlates with disease progression in patients with colorectal cancer. PLoS One 2013;8:e53630. [Crossref] [PubMed]
- Niu J, Jiang C, Li C, et al. Foxp3 expression in melanoma cells as a possible mechanism of resistance to immune destruction. Cancer Immunol Immunother 2011;60:1109-18. [Crossref] [PubMed]
- Chen DJ, Li XS, Zhao H, et al. Inhibitory effect of lentiviral-mediated RNA on the expression of Foxp3 protein in melanoma cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2012;28:337-9. [PubMed]
- Cunha LL, Morari EC, Nonogaki S, et al. Foxp3 expression is associated with aggressiveness in differentiated thyroid carcinomas. Clinics (Sao Paulo) 2012;67:483-8. [Crossref] [PubMed]
- Kakita N, Kanto T, Itose I, et al. Comparative analyses of regulatory T cell subsets in patients with hepatocellular carcinoma: a crucial role of CD25(-) FOXP3(-) T cells. Int J Cancer 2012;131:2573-83. [Crossref] [PubMed]
- Wei S, Kryczek I, Edwards RP, et al. Interleukin-2 administration alters the CD4+FOXP3+ T-cell pool and tumor trafficking in patients with ovarian carcinoma. Cancer Res 2007;67:7487-94. [Crossref] [PubMed]
- Lal G, Bromberg JS. Epigenetic mechanisms of regulation of Foxp3 expression. Blood 2009;114:3727-35. [Crossref] [PubMed]
- Bennett CL, Yoshioka R, Kiyosawa H, et al. X-Linked syndrome of polyendocrinopathy, immune dysfunction, and diarrhea maps to Xp11.23-Xq13.3. Am J Hum Genet 2000;66:461-8. [Crossref] [PubMed]
- Walker MR, Kasprowicz DJ, Gersuk VH, et al. Induction of FoxP3 and acquisition of T regulatory activity by stimulated human CD4+CD25- T cells. J Clin Invest 2003;112:1437-43. [Crossref] [PubMed]
- Zorn E, Kim HT, Lee SJ, et al. Reduced frequency of FOXP3+ CD4+CD25+ regulatory T cells in patients with chronic graft-versus-host disease. Blood 2005;106:2903-11. [Crossref] [PubMed]
- Lopez-Pastrana J, Shao Y, Chernaya V, et al. Epigenetic enzymes are the therapeutic targets for CD4(+)CD25(+/high)Foxp3(+) regulatory T cells. Transl Res 2015;165:221-40. [Crossref] [PubMed]
- Janson PC, Winerdal ME, Marits P, et al. FOXP3 promoter demethylation reveals the committed Treg population in humans. PLoS One 2008;3:e1612. [Crossref] [PubMed]
- Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature 2010;465:721-7. [Crossref] [PubMed]
- Ma AN, Wang H, Guo R, et al. Targeted gene suppression by inducing de novo DNA methylation in the gene promoter. Epigenetics Chromatin 2014;7:20. [Crossref] [PubMed]
- Doerfler W, Weisshaar B, Hoeveler A, et al. Promoter inhibition by DNA methylation: a reversible signal. Gene 1988;74:129-33. [Crossref] [PubMed]
- Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol 2010;28:1069-78. [Crossref] [PubMed]
- Pixberg CF, Raba K, Muller F, et al. Analysis of DNA methylation in single circulating tumor cells. Oncogene 2017;36:3223-31. [Crossref] [PubMed]
- Zhang YJ, Rossner P Jr, Chen Y, et al. Aflatoxin B1 and polycyclic aromatic hydrocarbon adducts, p53 mutations and p16 methylation in liver tissue and plasma of hepatocellular carcinoma patients. Int J Cancer 2006;119:985-91. [Crossref] [PubMed]
- Ying L, Yan F, Meng Q, et al. PD-L1 expression is a prognostic factor in subgroups of gastric cancer patients stratified according to their levels of CD8 and FOXP3 immune markers. Oncoimmunology 2018;7:e1433520. [Crossref] [PubMed]
- West NR, Kost SE, Martin SD, et al. Tumour-infiltrating FOXP3(+) lymphocytes are associated with cytotoxic immune responses and good clinical outcome in oestrogen receptor-negative breast cancer. Br J Cancer 2013;108:155-62. [Crossref] [PubMed]
- Yang S, Liu Y, Li MY, et al. FOXP3 promotes tumor growth and metastasis by activating Wnt/beta-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer 2017;16:124. [Crossref] [PubMed]
- Song JJ, Zhao SJ, Fang J, et al. Foxp3 overexpression in tumor cells predicts poor survival in oral squamous cell carcinoma. BMC Cancer 2016;16:530. [Crossref] [PubMed]
- Mu L, Yang C, Gao Q, et al. CD4+ and Perivascular Foxp3+ T Cells in Glioma Correlate with Angiogenesis and Tumor Progression. Front Immunol 2017;8:1451. [Crossref] [PubMed]
- Flammiger A, Weisbach L, Huland H, et al. High tissue density of FOXP3+ T cells is associated with clinical outcome in prostate cancer. Eur J Cancer 2013;49:1273-9. [Crossref] [PubMed]
- Heinze E, Baldwin S, Chan G, et al. Antibody-mediated FOXP3 protein therapy induces apoptosis in cancer cells in vitro and inhibits metastasis in vivo. Int J Oncol 2009;35:167-73. [PubMed]
- Floess S, Freyer J, Siewert C, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol 2007;5:e38. [Crossref] [PubMed]


