
PRC1 promotes cell proliferation and cell cycle progression by regulating p21/p27-pRB family molecules and FAK-paxillin pathway in non-small cell lung cancer
Introduction
Lung cancer is the leading causes of cancer death around the world, with projected 154,050 lung cancer deaths occurring in 2018, and an estimated 234,030 new cases of lung cancer being diagnosed (1). Although various strategies have been applied to improve the treatment regimen for lung cancer, the 5-year survival of lung cancer is 10–20% in most countries (2,3). At present, there is still a lack of understanding of the molecular mechanism in the carcinogenesis of non-small cell lung cancer (NSCLC), which is the most common subtype of lung cancer (4).
Evidence shows that precise diagnosis and therapy are beneficial for improving survival of patients with lung cancer (5). Thus, it is essential to identify the carcinogenesis of lung cancer and therefore to find promising therapeutic targets. Through gene expression profile chip data analysis, the establishment of molecular marker groups related to the occurrence and development of lung cancer can provide abundant information for treatment (6). Mutations of EGFR, KRAS, and ALK have been demonstrated to be associated with the carcinogenesis of lung cancer (7). Consequently, various first-line drugs have been manufactured to realize precise target therapy (8).
Via analyzing three groups of gene expression profile data of NSCLC patients (GSE42127, GSE11969, GSE8894) from the Gene Expression Omnibus (GEO) database (9), a group of differential genes related to the malignant phenotype and poor prognosis of NSCLC were found, including AURKA, PRC1, COL17A1, and CDKN3. A protein regulator of cytokinesis 1 (PRC1), belonging to the microtubule-associated proteins (MAPs) family, is located on chromosome 15 in humans (10). PRC1 was originally identified as a mitotic spindle-associated cyclin-dependent kinase (Cdk) substrate in vitro, which encodes a protein present at high levels during the S and G2/M phases of mitosis (11). PRC1 has been subsequently regarded as a microtubule binding and bundling protein that maintains the midzone of the mitotic spindle (12). Microtubules play essential roles in the cell cycle, along with cell trafficking, signaling transduction, and cell migration (13). Alterations in microtubule-associated proteins have been demonstrated in carcinogenesis (14).
Growing evidence indicates that PRC1 is overexpressed in some malignant tumors and plays an important role in tumor progression (15,16). However, the function and molecular mechanism of PRC1 in the carcinogenesis of NSCLC are still unclear. This study aimed to identify the function and molecular mechanism of PRC1 in NSCLC, and further provide therapeutic targets for NSCLC patients in clinical practice.
Methods
Bioinformatics analysis
Gene expression profile and clinical outcomes of 285 patients with stage I–III were obtained from three datasets of GEO (GSE42127, GSE11969, GSE8894) (Table S1). All data were screened based on flag filtering, fold change filtering, and expression level filtering, and then selected data were standardized by the LOWESS method. Differentially expressed genes (DEGs) were selected and presented by Student’s t-test, similar sample research, scatter plot, cluster/gene tree, and Venn diagram. The KEGG pathways of 35 differentially expressed key genes were analyzed online by DAVID. The DEG sets were analyzed based on the database of KEGG biological pathways (http://www.genome.jp/). Meanwhile, gene ontology (GO) annotation and functional cluster analysis were applied to screen DEGs which were related to the poor prognosis of lung cancer. TELiS online and GATHER online were used to predict binding sites of transcription factors in DEGs. STRING online analysis was used to predict the expression patterns of DEGs. Moreover, the key DEGs were verified by analyzing their expression levels in tissues from TCGA datasets, and performing quantitative real-time polymerase chain reaction (q-PCR) based on 36 tumor tissues and corresponding adjacent non-tumor tissues collected from our institution.
Table S1
| GEO datasets | Platform | Samples (test/control) | Status | Submission date |
|---|---|---|---|---|
| GSE8894 | GPL570 | 62 (29/33) | Recurrence/no recurrence | Nov 20, 2007 |
| GSE11969 | GPL7015 | 90 (58/32) | Metastasis/no metastasis | Apr 01, 2009 |
| GSE42127 | GPL6884 | 133 (49/84) | ACT + good prognosis/ACT + poor prognosis | Jan 30, 2013 |
ACT, adjuvant chemotherapy.
Cell culture
NSCLC cell lines A549 and NCL-H358 were purchased from Institutes for Biological Science, Chinese Academy of Science. A549 and NCL-H358 cells were cultured in RPMI-1640 containing 10% fetal bovine serum at 37 °C in a humidified incubator containing 5% CO2.
Clinical sample collection
Patients who met the following criteria were enrolled in our study: (I) pathologically diagnosed with NSCLC from 2015 to 2016 in Ningbo First Hospital; (II) no chemotherapy or radiation received before surgery. Tumor tissues and corresponding adjacent non-tumor tissues at a minimum distance of 5 cm from tumor tissue were collected. Clinical data were also obtained from medical records. This study was approved by the Ethics Committee of Ningbo First Hospital, and informed consent was signed by all participants.
RNA extraction and q-PCR
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA of cultured cells and tissue samples. Total RNA concentrations, purity, and integrity were also assessed. Reverse transcription was carried out to obtain cDNA by Primer Script RT-reagent Kit (TaKaRa), while q-PCR was performed by SYBR Green PCR master mix. GAPDH was considered as an internal reference for relative quantitative analysis (Table S2).
Table S2
| Gene | Forward primer | Reverse primer |
|---|---|---|
| PRC1 | 5'- GCTGAGATTGTGCGGTTA-3' | 5'- GCCTTCAACTCTTCTTCCA-3' |
| GAPDH | 5'-TGACTTCAACAGCGACACCCA-3' | 5'-CACCCTGTTGCTGTAGCCAAA-3' |
Western blot (WB)
After being transfected with highly expressed PRC1 plasmid or siRNA-PRC1, cultured cells were lysed by lysis buffer (RIPA, KeyGEN) with protease inhibitors (PMSF, KeyGEN) on ice for 30 minutes. A BCA Kit (KeyGEN) was used to determine protein concentration. Approximately 30 µg of lysate protein was used for WB. Polyvinylidene fluoride (PVDF) was scanned by ChemiDoc™ XRS (Bio-Rad), and β-actin or GAPDH was regarded as the candidate protein.
Plasmid construction and cell transfection
The plasmids with high PRC1 expression and an empty vector were provided by GenePharma, Shanghai, China. The plasmids with a high expression of PRC1, empty vector, siRNA-PRC1, and siRNA- negative control (NC) were each transfected into NSCLC cells via Opti-MEM mediums containing Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) and plasmid NDA. The transfection efficiency was assessed 48 hours after transfection. Q-PCR and WB were performed to determine the expression of PRC1 mRNA and PRC1 protein, respectively (Table S3).
Table S3
| Plasmid | Sense (5'-3') | Antisense (5'-3') |
|---|---|---|
| PRC1-Homo-1816 | GGAAAGCGCUGCAAUUAGATT | UCUAAUUGCAGCGCUUUCCTT |
| PRC1-Homo-1749 | CUGAGGUGGUAAAGAAGCATT | UGCUUCUUUACCACCUCAGTT |
| PRC1-Homo-852 | CUGUGCGAAAUUCUUUGUATT | UACAAAGAAUUUCGCACAGTT |
Soft agar assay
The 0.3% upper soft agar was configured with low melting agarose and 2×RPMI-1640 mediums (Corning, NY, USA). After adding 0.3% upper soft agar into 6-well plates containing 0.6% underlying soft agar, 100 uL single cell suspensions were seeded. After a 2–3 weeks culture, inverted fluorescence microscope, combined with the following formula, was used to evaluate cell proliferation: proliferation efficiency (PE) = (colony number/inoculated cell number)*100%.
Cell transwell
RPMI-1640 medium with 500 µL of 10% fetal bovine serum, 100 µL serum-free RPMI-1640, and 3×104 cells were added into the lower chamber of a transwell (Corning, NY, USA) for 12 hours at 37 °C. After culture, the cells on the upper surface of the membrane were wiped off, while the cells on the lower surface of the membrane were fixed with paraformaldehyde for 15 minutes and stained with crystal violet for 5 minutes. The cells fixed on the lower surface of the membrane were counted by microscope.
Flow cytometry
Negative control and NSCLC cells after transfection were digested and blown into cell suspension. After repeated PBS washing and centrifugation, RnaseA and PI were added to the PBS containing cells. Then, 500 uL of this mixture were stained in a cell incubator at 37 °C, and Beckman Coulter Fc500 was applied to analyze the distribution of cell cycle.
Immunohistochemistry
To deparaffinize and rehydrate tissue sections, 95%, 90%, 85%, 80%, and 70% alcohol were used, while 3% H2O2 was applied to block endogenous peroxidase activity. Antigen retrieval was performed by 5% sheep serum. After adding PRC1 antibodies (Abcam), diamino benzidine (DAB) was used for immunostaining. A microscope was used to evaluate staining intensity and the percentage of positive staining cells, while Olympus imaging system was applied to quantitatively analyze the staining results of PRC1 protein. The mean optical density (MOD) was used to assess the expression level of PRC1.
Animal experiments
All animal experiments were performed based on the NIH animal use guidelines and approved by the Ethics Committee of Ningbo First Hospital. The plasmids with a high-expression of PRC1 and empty vector were each transfected into tumor cells. Then, 10 mice (Laboratory Animal Resources, Chinese Academy of Science) were used for tumor transplantation, and 200 uL cell suspension was injected into each selected mouse aged 4–6 weeks and weighing 18–22 kg. The volume of tumor was calculated by the length and weight of the tumor. The growth curve of the tumor was also plotted. Tumor tissues were obtained about 5 weeks after transplantation.
Cell immunofluorescence assay
After being transfected for 72 hours, cells were digested and suspended. After fixation and washing by PBS, fluorescent antibodies (Santa Cruz Biotechnology) were added and reacted with antigens. An inverted fluorescence microscope was used to assess the fluorescence signal of each group. Image J software was used to analyze immunofluorescence intensity (IOD).
Cell counting kit-8 (CCK-8)
All tumor cells were plated into 96-well plates and each well contained 100 uL culture medium and 5×103 cells. All tumor cells after transfection were divided into 4 groups: the plasmids with a high-expression of PRC1, empty vector, PRC1-siRNA-negative control, and PRC1-siRNA-FAK. Each well was infused with 10 uL CCK-8 regent (Dojindo, Japan) after transfection for 48 hours. Then, all tumor cells were incubated at 37 °C for 2 hours. When the color of the culture medium became orange, ELISA (Bio-Rad) was used to detect absorbance at 450 mm to evaluate the impact of PRC1 on the proliferation of NSCLC cells and the mechanisms regulated by the FAK pathway.
Statistical analysis
All continuous data are expressed as mean ± standard. Student’s t-test was conducted to analyze any differences between the two groups. Kaplan-Meier and log-rank tests were performed to analyze the relationship between the expression level of PRC1 and the survival of patients. P<0.05 was marked as *, indicating statistical difference; while P<0.01 was marked as **, indicating statistically significant difference.
Results
PRC1 may be a potential oncogene of NSCLC
Altogether, 35 DEGs including 29 upregulated genes were identified by GSE42127, GSE11969, and GSE8894. Most of the upregulated genes were associated with cell cycle, extracellular matrix receptor regulation, and cell movement. After reviewing other studies (17,18), PRC1, which might be related to the poor prognosis of NSCLC, was selected as our target. The TCGA dataset indicated that the expression level of PRC1 in tumor tissue was higher than that of adjacent non-tumor tissues in NSCLC (P<0.01) (Figure 1A). The characteristics of the 36 enrolled patients are shown in Table 1. Q-PCR reconfirmed that the higher expression of PRC1 mRNA in tumor tissue in NSCLC (P<0.01) (Figure 1B).

Table 1
| Patient parameters (n=36) | Number |
|---|---|
| Sex (male/female) | 20/16 |
| Age (years old) | |
| >60 | 21 |
| ≤60 | 15 |
| Smoking status (smoking/non-smoking) | 19/17 |
| Maximum diameter of primary tumor | |
| ≤3 cm | 23 |
| >3 cm | 13 |
| Lymphatic metastasis (+/−) | 12/24 |
| TNM stage (I/II/III/IV) | 13/15/8/0 |
| Subtypes of lung cancer | |
| Squamous cell carcinoma | 14 |
| Adenocarcinoma | 21 |
| Large cell lung cancer | 1 |
PRC1 overexpression promoted NSCLC cell proliferation and migration, and accelerated the process of cell cycle in vitro
Cells transfected with highly expressed PRC1 plasmids, empty vector, siRNA-PRC1, and siRNA-NC, were divided into the TOPO-PRC1 group, TOPO-Vector group, siRNA-PRC1 group, and siRNA-NC group, respectively. Compared with the TOPO-Vector group, PRC1 protein and mRNA expression of cells were upregulated significantly in the TOPO-PRC1 group (P<0.01) (Figure 2A). Compared with the siRNA-NC group, cells in the siRNA-PRC1 group had down-regulated PRC1 protein and mRNA expression (P<0.01) (Figure 2B). Soft agar assay showed that cells in TOPO-PRC1 group had significantly higher PE than those in the control group, while lower PE appeared in the siRNA-PRC1 group, compared with the TOPO-Vector group (P<0.05) (Figure 2C,D).
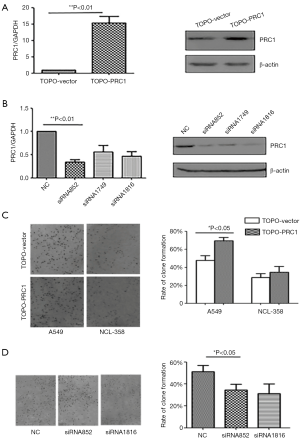
PRC1 expression level was related to NSCLC cell cycle and its migration
Flow cytometry indicated that compared with TOPO-PRC1 group, TOPO-Vector group had more cells in the G2/M phase (8%±0.53% vs. 12%±0.75%) and less cells in the S phase, with the differences being significant (P<0.05) (Figure 3A,B). Cell transwell showed that compared with the other 3 groups, more NSCLC cells in the TOPO-PRC1 group migrated to the lower surface of the membrane (P<0.05) (Figure 3C,D).
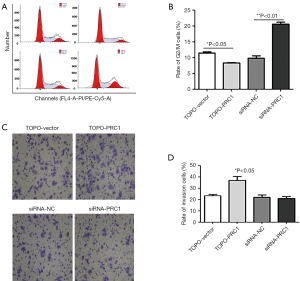
The upregulation of PRC1 expression promoted tumor growth in mice
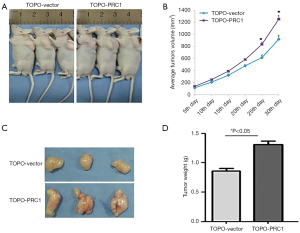
No mice died before the removal of tumor tissues, and tumors were obtained from all mice. Tumors in the mice of the TOPO-PRC1 group grew faster, and from the third week of transplantation, the difference of tumor size gradually became great (P<0.05). The TOPO-PRC1 group had larger tumor sizes and significantly heavier tumor weight than the TOPO-Vector group (Figure 4).
The high expression of PRC1 was associated with late stage and poor prognosis for NSCLC patients
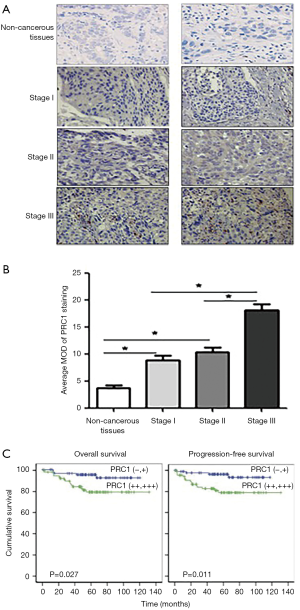
A total of 150 eligible NSCLC patients were included to assess the relationship between PRC1 expression and survival of NSCLC patients. For 150 included patients, the 1-year follow-up rate, 3-year follow-up rate, and 5-year follow-up rate were 100%, 83.6%, and 61.2%, respectively. Only 1 patient was lost to follow-up after 3 years. The basic characteristics of the 150 patients are shown in Table 2. The immunohistochemical results not only indicated that PRC1 protein was mainly located in the cytoplasm and partially in the nucleus, but also suggested that patients with different stages had different staining intensity (Figure 5A). For stage III NSCLC patients, the tumor tissue had significantly higher MOD than that of patients with other stages, and the MOD of tumor tissue was higher than that of adjacent non-tumor tissue in stage III patients, but not for patients with other stages (Figure 5B). In addition, higher PRC1 expression was also related to lymph node metastasis and adenocarcinoma (P<0.05). Survival analysis implied a significantly meaningful trend towards shorter overall survival and progression free survival with the higher PRC1 expression (P<0.05) (Figure 5C).
Table 2
| Characteristics | Number | Number of PRC1 positive expression | MOD (mean ± SD) | P |
|---|---|---|---|---|
| Sex | 0.221 | |||
| Male | 91 | 78 | 12.56±4.43 | |
| Female | 59 | 52 | 10.69±3.77 | |
| Age (years old) | 0.137 | |||
| ≤60 | 87 | 74 | 10.73±3.65 | |
| >60 | 63 | 56 | 12.78±5.94 | |
| Lymphatic metastasis | <0.001 | |||
| + | 56 | 56 | 18.05±2.03 | |
| − | 94 | 74 | 9.53±3.72 | |
| TNM stage | <0.001 | |||
| I | 52 | 35 | 8.89±1.42 | |
| II | 42 | 39 | 10.33±1.56 | |
| III | 56 | 56 | 18.05±2.03 | |
| IV | 0 | 0 | Non-available | |
| Subtype of lung cancer | <0.001 | |||
| Squamous cell carcinoma | 38 | 33 | 9.11±2.71 | |
| Adenocarcinoma | 110 | 97 | 14.29±4.59 | |
| Large cell lung cancer | 2 | 0 | Non-available |
MOD, mean optical density.
PRC1 promoted NSCLC proliferation by regulating the phosphorylation of molecules in the FAK-Paxillin pathway
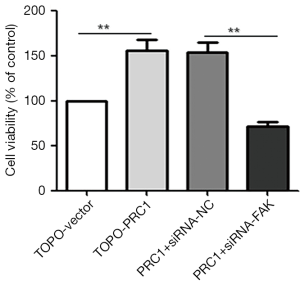
All experiments of CCK-8 showed that the NSCLC cells in the TOPO-PRC1 group had a higher ability of proliferation compared with that in the TOPO-Vector group (**P<0.01). Meanwhile, the proliferation capacity of NSCLC cells in the PRC1+siRNA-FAK group was significantly lower than that in the PRC1+siRNA-NC group (**P<0.01) (Figure 6).
WB was performed to detect the phosphorylation level of several major phosphorylation sites at the N-terminal, C-terminal, and the kinase activation ring of NSCLC cells in the 4 groups. In the TOPO-PRC1 group, the FAK pY397, FAK Py576/577, and FAK pY925 of NSCLC cells showed a significantly higher phosphorylation level than those cells in the TOPO-vector group, which was in line with the quantitative analysis of the Gel pro analyzer 4.0 (*P<0.05) (Figure 7A). Moreover, the Gel pro analyzer 4.0 also showed that the difference in the phosphorylation level of FAK pY576/577 between the TOPO-PRC1 group and TOPO-Vector group was greatest (**P<0.01) (Figure 7B). The expression of FAK pY397, FAK pY576/577, and FAK pY925 significantly decreased in the siRNA-PRC1 group (**P<0.01) (Figure 7C).
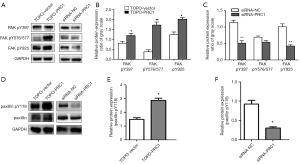
WB also implied that the phosphorylation level of paxillin, the downstream target of FAK signaling pathway, was upregulated in the TOPO-PRC1 group (Figure 7D), while the phosphorylation level of paxillin was down-regulated in the siRNA-PRC1 group (*P<0.05) when compared with that of the TOPO-Vector group and siRNA-NC group, respectively (Figure 7E,F).
PRC1 influenced the NSCLC cell cycle by regulating the phosphorylation of p21/p27 and pRB family molecules
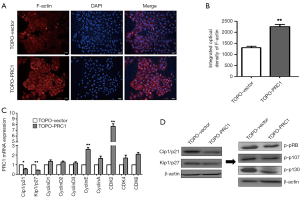
Immunofluorescence showed that the TOPO-PRC1 group had a higher IOD of F-actin than that of the TOPO-Vector group (Figure 8A,B). Compared with those in the TOPO-Vector group, cells in the TOPO-PRC1 group had lower expression of Cip1/p21 mRNA (*P<0.05) and Kip1/p27 mRNA (**P<0.01). Meanwhile, WB indicated that the TOPO-PRC1 group also had higher expressions of both Cip1/p21 and Kip1/p27 protein (Figure 8C). Furthermore, NSCLC cells in the TOPO-PRC1 group showed lower phosphorylation levels of p107 and p130 than those in the TOPO-Vector group, while no difference was found in the phosphorylation levels of pRB between these 2 groups (Figure 8D).
Discussion
Our trials showed a high expression of PRC1 could promote the proliferation and migration of NSCLC cells, and the growth of NSCLC xenografts in mice. These results strongly confirm that the upregulation of PRC1 expression may play an important role in the occurrence and development of NSCLC, as evinced by the cell models, animal models, and clinical samples.
PRC1 accelerated proliferation of NSCLC cells through FAK signaling pathway
FAK is a cytoplasmic non-receptor protein tyrosine kinase and cytoskeleton protein, involved in p53, PI3K, and other cell signaling pathways by phosphorylation (19). FAK phosphorylation affects cell proliferation, migration, and invasion (20), and its roles in tumorigenesis of some solid tumors has been confirmed by previous research (21). In this study, we found inhibiting FAK signaling pathway could inhibit the promoting effect of PRC1 on the proliferation of NSCLC cells, suggesting that PRC1 may affect the proliferation of NSCLC cells by regulating FAK signaling pathway. We also verified the positive relationship between PRC1 expression in NSCLC cells and the phosphorylation level of the main phosphorylation sites in different functional regions of FAK (FAK pY397, FAK pY576/577, and FAK pY925). Thus, PRC1 might accelerate NCLC cell proliferation by promoting the phosphorylation level of multiple phosphorylation sites in FAK signaling pathway.
FRC1 promotes proliferation through FAK/paxillin signaling pathway
Our study showed that PRC1 regulated the phosphorylation level of both FAK and paxillin pY118 by affecting the phosphorylation level of paxillin, which is in line with two previous studies (22,23). FAK/paxillin signaling pathway is closely related to the proliferation, adhesion, invasion, and metastasis of a variety of tumor cells, and therefore participates in the tumorigenesis, development, and metastasis of cancer (24). Clearly, PRC1 can promote the proliferation of NSCLC cells by regulating the phosphorylation level of the molecules in FAK-paxillin pathway.
Relationship with cell cycle: F-actin, Cip1/p21, Kip1/p27, and pRb family
Our trial indicated that PRC1 affected the cell cycle of tumor cells by regulating F-actin, thus promoting cell proliferation and migration. It has been reported that F-actin is a spiral fibrous polymer composed of globular actin subunits (25). F-actin plays an important role in cell adhesion, migration, apoptosis, and promotion of the invasive growth of cells (26). Current investigation implies that F-actin can regulate cytoskeleton components through integrin and FAK signal transduction pathways (27).
The cell cycle is completed through the interaction between cyclin and cyclin dependent kinase (CDK) (28). Cip1/p21 and Kip1/p27 are the negative regulators of the cell cycle and members of the cyclin-dependent kinase inhibitor (CDKI) family, and are able to inhibit the kinase activity of a variety of cyclin and almost all cyclin-CDK complexes, therefore playing an anti-tumor role (29). This study showed that Cip1/p21 and Kip1/p27 expressions were significantly inhibited in NSCLC cells with a high expression of PRC1, consistent with the findings of two previous studies that show low expressions of Kip1/p27 being associated with poor prognoses (30,31). Therefore, Kip1/p27 was a tumor suppressor gene and could be used as an independent prognostic indicator for malignant tumors.
Cip 1/p21 is the most widely known cell cycle inhibitor protein with the most extensive inhibitory kinase activity (32). Cip 1/p21 widely inhibits the phosphorylated kinase activity of cyclin-CDK complexes in G1 and S phases (32). The negative regulation of the cell cycle is realized by two main mechanisms: (I) the binding of Cip1/p21 with cyclin-CDK complex inhibiting the phosphorylation of pRb, and thus stagnating the cell cycle; (II) the binding of Cip 1/p21 with proliferating cell nuclear antigen forming a complex to inhibit DNA replication and stagnate the cell cycle in the Gl phase (33). A similar function is also found between Cip1/p21 and Kip1/p27, but a lower inhibitory effect on the tumor cell cycle is present in Cip1/p21 than in Kip1/p27 (34). Given this, the deletion of Cip1/p21 in tumor may be a promising prognostic indicator.
Strengths and limitations
This study verified the role of PRC1 in the development of NSCLC based on both cell models and animal models. In addition, the impact of PRC1 on the prognosis of patients strongly indicated us that the therapy targeted to PRC1 is a promising regimen to improve the survival of NSCLC patients. Nevertheless, this work inevitably had some limitations. First, the limited sample size restricted us to exclude the possibility that some lung cancer cells mainly follow other alternative signaling pathways related to PRC1. Second, the relationships between FAK-paxillin and cell proliferation, as well as p21/p27 or pRb and cell cycle, were demonstrated by analyzing some published researches but not in this experiment. Third, this study did not investigate the effect of epigenetic modifications related to PRC1. These limitations will be further solved in our future work.
Conclusions
PRC1 was significantly upregulated in tumor tissue and promoted tumor cell proliferation, migration, and cell cycle progression in NSCLC. The effects of PRC1 were achieved by regulating the phosphorylation level of p21/p27-pRB family molecules and FAK-paxillin signaling pathway molecules, thus promoting NSCLC cell proliferation. This study indicates that PRC1 is a key therapeutic target for NSCLC patients, and this discovery can aid in the development of further exploration of the molecular mechanism of PRC1 in NSCLC.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.19). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Ningbo First Hospital, and informed consent was signed by all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 2015;385:977-1010. [Crossref] [PubMed]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Gao D, Vahdat LT, Wong S, et al. Microenvironmental regulation of epithelial-mesenchymal transitions in cancer. Cancer Res 2012;72:4883-9. [Crossref] [PubMed]
- Wang DC, Wang W, Zhu B, et al. Lung Cancer Heterogeneity and New Strategies for Drug Therapy. Annu Rev Pharmacol Toxicol 2018;58:531-46. [Crossref] [PubMed]
- Liu J, Lee W, Jiang Z, et al. Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events. Genome Res 2012;22:2315-27. [Crossref] [PubMed]
- Warth A, Penzel R, Lindenmaier H, et al. EGFR, KRAS, BRAF and ALK gene alterations in lung adenocarcinomas: patient outcome, interplay with morphology and immunophenotype. Eur Respir J 2014;43:872-83. [Crossref] [PubMed]
- Lopez-Chavez A, Thomas A, Rajan A, et al. Molecular profiling and targeted therapy for advanced thoracic malignancies: a biomarker-derived, multiarm, multihistology phase II basket trial. J Clin Oncol 2015;33:1000-7. [Crossref] [PubMed]
- Tang H, Xiao G, Behrens C, et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res 2013;19:1577-86. [Crossref] [PubMed]
- Shrestha S, Wilmeth LJ, Eyer J, et al. PRC1 controls spindle polarization and recruitment of cytokinetic factors during monopolar cytokinesis. Mol Biol Cell 2012;23:1196-207. [Crossref] [PubMed]
- Kurasawa Y, Earnshaw WC, Mochizuki Y, et al. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J 2004;23:3237-48. [Crossref] [PubMed]
- Mollinari C, Kleman JP, Saoudi Y, et al. Ablation of PRC1 by small interfering RNA demonstrates that cytokinetic abscission requires a central spindle bundle in mammalian cells, whereas completion of furrowing does not. Mol Biol Cell 2005;16:1043-55. [Crossref] [PubMed]
- Ganguly A, Yang H, Sharma R, et al. The role of microtubules and their dynamics in cell migration. J Biol Chem 2012;287:43359-69. [Crossref] [PubMed]
- Cirillo L, Gotta M, Meraldi P. The Elephant in the Room: The Role of Microtubules in Cancer. Adv Exp Med Biol 2017;1002:93-124. [Crossref] [PubMed]
- Luo HW, Chen QB, Wan YP, et al. Protein regulator of cytokinesis 1 overexpression predicts biochemical recurrence in men with prostate cancer. Biomed Pharmacother 2016;78:116-20. [Crossref] [PubMed]
- Chen J, Rajasekaran M, Xia H, et al. The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinoma in association with the Wnt/beta-catenin signalling pathway. Gut 2016;65:1522-34. [Crossref] [PubMed]
- Zhan P, Xi GM, Liu HB, et al. Protein regulator of cytokinesis-1 expression: prognostic value in lung squamous cell carcinoma patients. J Thorac Dis 2017;9:2054-60. [Crossref] [PubMed]
- Zhan P, Zhang B, Xi GM, et al. PRC1 contributes to tumorigenesis of lung adenocarcinoma in association with the Wnt/beta-catenin signaling pathway. Mol Cancer 2017;16:108. [Crossref] [PubMed]
- Kleinschmidt EG, Schlaepfer DD. Focal adhesion kinase signaling in unexpected places. Curr Opin Cell Biol 2017;45:24-30. [Crossref] [PubMed]
- Zhao X, Guan JL. Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv Drug Deliv Rev 2011;63:610-5. [Crossref] [PubMed]
- Chuanyu S, Yuqing Z, Chong X, et al. Periostin promotes migration and invasion of renal cell carcinoma through the integrin/focal adhesion kinase/c-Jun N-terminal kinase pathway. Tumour Biol 2017;39:1010428317694549. [Crossref] [PubMed]
- Miller NL, Lawson C, Kleinschmidt EG, et al. A non-canonical role for Rgnef in promoting integrin-stimulated focal adhesion kinase activation. J Cell Sci 2013;126:5074-85. [Crossref] [PubMed]
- Sachdev S, Bu Y, Gelman IH. Paxillin-Y118 phosphorylation contributes to the control of Src-induced anchorage-independent growth by FAK and adhesion. BMC Cancer 2009;9:12. [Crossref] [PubMed]
- Chen JY, Tang YA, Huang SM, et al. A novel sialyltransferase inhibitor suppresses FAK/paxillin signaling and cancer angiogenesis and metastasis pathways. Cancer Res 2011;71:473-83. [Crossref] [PubMed]
- Maruthamuthu V, Aratyn-Schaus Y, Gardel ML. Conserved F-actin dynamics and force transmission at cell adhesions. Curr Opin Cell Biol 2010;22:583-8. [Crossref] [PubMed]
- Stricker J, Falzone T, Gardel ML. Mechanics of the F-actin cytoskeleton. J Biomech 2010;43:9-14. [Crossref] [PubMed]
- Ni Y, Wang X, Yin X, et al. Plectin protects podocytes from adriamycin-induced apoptosis and F-actin cytoskeletal disruption through the integrin alpha6beta4/FAK/p38 MAPK pathway. J Cell Mol Med 2018;22:5450-67. [Crossref] [PubMed]
- Santo L, Siu KT, Raje N. Targeting Cyclin-Dependent Kinases and Cell Cycle Progression in Human Cancers. Semin Oncol 2015;42:788-800. [Crossref] [PubMed]
- Whittaker SR, Mallinger A, Workman P, et al. Inhibitors of cyclin-dependent kinases as cancer therapeutics. Pharmacol Ther 2017;173:83-105. [Crossref] [PubMed]
- Guan X, Wang Y, Xie R, et al. p27(Kip1) as a prognostic factor in breast cancer: a systematic review and meta-analysis. J Cell Mol Med 2010;14:944-53. [Crossref] [PubMed]
- Zhuang Y, Yin HT, Yin XL, et al. High p27 expression is associated with a better prognosis in East Asian non-small cell lung cancer patients. Clin Chim Acta 2011;412:2228-31. [Crossref] [PubMed]
- Kreis NN, Louwen F, Yuan J. Less understood issues: p21(Cip1) in mitosis and its therapeutic potential. Oncogene 2015;34:1758-67. [Crossref] [PubMed]
- Yoon MK, Mitrea DM, Ou L, et al. Cell cycle regulation by the intrinsically disordered proteins p21 and p27. Biochem Soc Trans 2012;40:981-8. [Crossref] [PubMed]
- Abukhdeir AM, Park BH. P21 and p27: roles in carcinogenesis and drug resistance. Expert Rev Mol Med 2008;10:e19. [Crossref] [PubMed]


