
MicroRNA-204-3p modulates epithelial-mesenchymal transition by targeting paired box gene 2 in human melanoma A-375 cells
Introduction
Cutaneous melanoma, also known as malignant melanoma, is the most malignant type of skin cancer (1). Melanocytes emanate from neural crest cells and responsible for the production of pigment melanin. Numerous factors, especially under UV exposure, promote normal melanocytes to transform into malignant melanomas (2). Although the incidence of melanoma is lower than other skin cancers, melanoma is the main cause of skin cancer-related death with over 75% 5-year mortality rate (3). More worryingly, the incidence of melanoma has been rising steadily in the past 30 years, even in those countries which recorded a lower risk of melanoma in history (4). In case that primary melanoma is diagnosed and surgical resection early, most patients with melanomas can be cured. However, there are still a large proportion of patients facing severe risk of metastasis. Numerous researchers revealed that metastasis was a complex process and was closely implicated in poor prognosis and mortality of many cancers (5,6). Animal experiments showed that less than 0.01% cancer cells eventually developed into metastatic cancers after entering the circulation (7,8). However, malignant melanoma has been found in lymphoid nodule, liver, lung and brain metastases in early-stage clinical trials (9), and the plastic, highly migratory behavior of melanoma created their rapid metastatic abilities (10). Therefore, it is necessary to further understand the molecular mechanism of melanoma metastasis contributing to the discovery of new biomarkers in order to effectively target melanoma metastasis and provide novel approaches to the prevention of cancer dissemination.
Paired box gene 2 (PAX2) is a member of paired box family, and the nuclear transcription factor encoded by PAX2 is essential for the differentiation of the renal tubular epithelial cell in the early stages of embryonic development (11,12). In general, the expression of PAX2 will disappear after the nephron is mature. However, high level PAX2 has been detected in various tumor cells such as renal tumor (13), ovarian cancer (14) and breast cancer (15). Previous researches showed that the upregulated PAX2 promoted epithelial-mesenchymal transition (EMT) and induced the process of renal interstitial fibrosis (16). More importantly, the relation between the migration and invasion abilities of melanoma cells and the ectopic expression of PAX2 has been also proved. PAX2 knockdown contributed to abolishing the resistant to cisplatin of melanoma cells (17). However, the molecular mechanism of PAX2 acting on the EMT process of melanoma cells remains relatively poorly understood.
MicroRNAs, small noncoding RNAs (18–25 nt), extensively participate in regulating the expressions of genes through targeting the 3'-untranslated region (UTR) of their mRNAs (18). At present, miRNAs were used as biomarkers for diagnosing various types of cancers including melanoma (19). MiR-204 was believed to act as an effective inhibitor in tumorigenesis, and low level miR-204 could strengthen the capacity of migration and invasion (20). The miR-204-5p and miR-204-3p were two types of mature miRNAs of miR-204, and miR-204-5p has been proved to act as a tumor inhibitor in malignant melanoma development (21). However, the function of miR-204-3p is still unclear. This study was performed to further verify the potential role of miR-204-3p in malignant melanoma.
Methods
Cell culture
Human immortalized epidermal cell line (HaCaT, #BNCC101683) was obtained from BeNa culture collection. A total of five human melanoma cell lines used in this study, including 451Lu cell line, A-375, VMM5A, SK-MEL-1 and G-361 cell lines. Only the first cell line was kindly provided by Dr. MeenhardHerlyn (Wistar Institute, PA, USA), the rest four ones were all purchased from the American Type Culture Collection (#CRL-1619, #CRL-3226, #HTB-67 and #CRL-1424™). HaCaT cells were cultured with Minimum Essential Medium (MEM, #56419C, Sigma-Aldrich, St. Louis, MO, USA), and all melanoma cell lines were incubated with Dulbecco’s modified eagle’s medium (DMEM, #D5030, Sigma-Aldrich). Both cell culture mediums contained 10% fetal bovine serum (FBS, #10099141, ThermoScientific, Hudson, NH, USA) and 1% penicillin-streptomycin, and then the cells were kept in a humidified incubator with 5% CO2 at 37 °C.
MiR-204-3p overexpression/inhibition
The sequence of MiR-204-3p was obtained from miRbase (accession number: MIMAT0022693). The cells were embedded into 12-well plates (1×105 cells per well) until the confluence reached 50%. MiR-204-3p mimics (50 nM), inhibitor (30 nM), negative control (50 nM), and inhibitor control (IC, 30 nM) were mixed into 250 µL DMEM medium, respectively. After that, Lipo-fectamine 2000 (Invitrogen, Carlsbad, CA, USA) was added into the DMEM medium. The mixture of diluted vector and transfection reagents were cultured at 37 °C in 5% CO2 humidified atmosphere for 6 h, and then continued to be cultured with complete medium. After 24 h, A-375 cells were harvested and qRT-PCR was used to determine the efficacies of the miR-203-3p mimic vector.
Cell viability detection
The viability of A-375 cells was determined using CCK-8 kit (Dojindo, Kumamoto, Japan) following the manufacturer’s instructions. The cells were embedded into 96-well plates and diluted to 3×105 cells/well in a humidified incubator with 5% CO2 at 37 °C. At 0, 24 and 48 h, medium was replaced by complete cell culture medium containing 10% CCK-8 reagent and then incubated about 4 h at room temperature. Each groups’ absorbance at 450 nm was measured by a Microplate Reader (Multiskan FC, Thermo Scientific).
Wound healing assay
The effects of miR-204-3p overexpression/inhibition on the migration of A-375 cells were investigated by wound healing assay. The cells were incubated in 6-well plates (5×104 cells/well) until the confluence reached 90%. Following culturing with complete medium for 24 h, a sterile pipette tip was used to produce a linear wound. Cell migration rate was calculated by comparing distance displaced by the cells at the acellular front 24 h later. Images were acquired using a light microscopy (Nikon, Tokyo, Japan).
Transwell assay
The capacity of A-375 cell invasion was examined using Transwell chamber assay. 1×105 cells were resuspended with serum-free DMEM and incubated into the upper chamber with polycarbonate membranes (8-mm pore-size, Corning, USA) lidded with matrigel (BD Biosciences, San Jose, CA, USA). The bottom chamber containing DMEM medium with 10% FBS was kept for 24 h at 37 °C. The cells on the upper surface of the membrane were wiped by swabbing. The cells that invaded onto the lower surface were fixed with methanol for 30 min and stained with 5% crystal violet solution. Three field views were randomly counted and averaged.
MiR-204-3p target prediction and luciferase reporter assay
We compared different prediction algorithms (TargetScan: http://www.targetscan.org/vert_72/ and miRTarBase: http://mirtarbase.mbc.nctu.edu.tw/) to reveal that the PAX2 and guanine nucleotide binding protein (G protein) q polypeptide (GNAQ) genes were potential targets of miR-204-3p. We cloned miR-204-3p into GV272 plasmid (GV272- miR-204-3p) and inserted the fragments containing miR-204-3p-binding site in the 3'UTR of PAX2 (accession number NM_001110.2) and GNAQ (accession number NM_002072) into GV268 plasmid (GV268-PAX2-3'UTR and GV268-GNAQ-3'UTR). The luciferase plasmids were obtained from Shanghai Genechem Co., Ltd. The QuikChangeH Site-Directed Mutagenesis Kit (Santa Clara, CA, USA) was used to destroy binding, and similarly the mutation fragments was installed into GV268 plasmid (GV268-PAX2-3'UTR mut and GV268-GNAQ-3'UTR mut).
Luciferase reporter assay was shown in the human kidney epithelial cell line (HEK293T, #CRL-3216, ATCC) by the dual-luciferase assay system (E1910; Promega, Madison, WI, USA). HEK293T Cells were co-transfected with GV272-miR-204-3p and GV268-PAX2-3'UTR (or GV268-PAX2-3'UTR mut) using Lipo-fectamine 2000 (Invitrogen) following the manufacturer’s instructions. After 48 h, HKR239T cells were lysed and separated by centrifugation at 15,000 ×g for 3 min. The dual luciferase reporter mixture was prepared to be mixed with the supernatant of lysed cells. The relative luciferase activity should be immediately measured by Sinergy 2 luminometer (Biotek, Winooski, VT). Firefly and Renilla luciferase activities of GNAQ gene were detected by the same method.
Cell apoptosis detection
PAX2 was identified as the potential target for miR-204-3p. siPAX2 was synthesized by Shanghai GenePharma Co., LTD. (Shanghai, China). To further confirm the role of PAX2 in the miR-204-3p-induced suppressive effects on the malignant melanoma, PAX2 siRNA was co-transfected with miR-204-3p inhibitor into A-375cells, and then detecting the changes in cell apoptosis.
For the determination of apoptosis rate, FITC Annexin V/PI apoptosis detection kit (Invitrogen) was performed on a flow cytometer (Becton Dickerson, CA, USA). The transfected cells were resuspended in 1× annexin-binding buffer and then incubated with FITC-conjugated Annexin V and propidium iodide (PI) (5 mg/mL) for 30 min. Apoptosis rates were analyzed by flow cytometry.
Quantitative real-time polymerase chain (qRT-PCR)
According to the manufacturer’s instructions, miRNAs were extracted from all samples by a miRNeasy Mini kit (Qiagen, Hilden, Germany), and RNAs were also manually extracted with TRIzol (Invitrogen, Carlsbad, CA, USA) and purified by TURBO DNA-free kit (Invitrogen). Firstly, miRNA was reversely transcribed using a miScript II RT Kit (Qiagen GmbH, Hilden, Germany), as described previously (22). The samples were reacted at 37 °C for 60 min, at 95 °C for 5 min and held at 4 °C. The relative miRNA expression was determined by the miScript SYBR Green PCR kit (Qiagen GmbH). Each qPCR was performed in a final volume of 20 µL containing 1× QuantiTect SYBR Green PCR Master mix (Qiagen, Hilden, Germany), 2 µL cDNA and 0.5 mM of each primer. The primer sequence of miR-204-3p was as follows: hsa-miR-204-3p forward: 5'-GCTGGGAAGGCAAAGGGACGT-3', and the miScript Universal primer was used as reverse primers. Secondly, prime script RT reagent kit (Takara, Shiga, Japan) was used to reversely transcribe extracted RNAs to cDNA. The reaction condition was as follow: first step at 65 °C for 5 min, the second was at 30 °C for 6 min and 50 °C for 60 min. qRT-PCR was performed using the SYBR green detection (Takara). Both experiments were analyzed by the ABI 7500 real-time PCR system. The reaction mixtures were incubated at 95 °C for 15 min, followed by 40 cycles of annealing at 94 °C for 15 sec, followed by at 55 °C for 30 sec and at 70 °C for 30 sec. All samples were carried out in triplicate, and the relative expression was calculated with the formula 2−ΔΔCt and normalized against U6 and GAPDH. All primers were shown in Table 1.
Table 1
| Gene name | Primer sequences |
|---|---|
| PAX2 | Forward: 5'-CCTGGCCACACCATTGTTC-3' |
| Reverse: 5'-TCACGTTTCCTCTTCTCACCAT-3' | |
| GNAQ | Forward: 5'-GACACCATCCTCCAGTTGAACC-3' |
| Reverse: 5'-ACACGCTCACACAGAGTCCAG-3' | |
| Hey-1 | Forward: 5'-GCCGACGAGACCGAATCAAT-3' |
| Reverse: 5'-GGAGACCAGGCGAACACGA-3' | |
| E-cadherin | Forward: 5'-GCCCTGCCAATCCCGATGAAA-3' |
| Reverse: 5'-GGGGTCAGTATCAGCCGCT-3' | |
| Vimentin | Forward: 5'-GCTTCAGAGAGAGGAAGCCGAAAA-3' |
| Reverse: 5'-CCGTGAGGTCAGGCTTGGAAA-3' | |
| MMP-2 | Forward: 5'-GGAGGCACGATTGGTCTG-3' |
| Reverse: 5'-TTGGTTTCCGCATGGTCT-3' | |
| MMP-9 | Forward: 5'-TTGACAGCGACAAGAAGTGG-3' |
| Reverse: 5'-GGCACAGTAGTGGCCGTAG-3' | |
| U6 | Forward: 5'-CTCGCTTCGGCAGCACA-3' |
| Reverse: 5'-AACGCTTCACGAATTTGCGT-3' |
qRT-PCR, quantitative real-time polymerase chain.
Western blot
Cells were lysed and collected by being centrifuged at 12,000 ×g for 20 min at 4 °C. The sample proteins were isolated by BCA Protein Assay Kit (Takara). Twenty µg proteins were subjected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad, Hercules, CA, USA), which were blocked with 5% skimmed milk/TBST for 1.5 h at 37 °C before being incubated with primary antibodies. The primary antibodies against PAX2 (c#9666), GNAQ (1:1,000, #3992) and several EMT-related genes such as epithelial cadherin (E-cadherin, 1:1,000, #14472), vimentin (1:1,000, #12826), matrix metalloproteinase 2 (MMP2) (1:1,000, #4022) and MMP-9 (1:1,000, #13667) were purchased from Cell Signaling Technology (Shanghai, China). The membranes were cultures with secondary antibodies (#7074 and #7076, 1:2,000, CST) for 1 h at room temperature. The protein level was analyzed using an ECL kit (32106, thermo Co. Ltd, USA) and normalized to GAPDH expression.
Statistical analysis
All results were presented as the mean ± SEM. The statistical significance of the studies was analyzed using one-way ANOVA. P<0.05 was considered as a statistically significant difference.
Results
MiR-204-3p overexpression inhibited the A-375 cell viability.
The results of qRT-PCR showed that the miR-204-3p expressions in all malignant melanoma cell lines (A-375, VMM5A, SK-MEL-1, 451Lu and G-361) were significantly lower than those in HaCaT cells (P<0.01, Figure 1A). As the miR-204-3p level in A-375 cells was the lowest, A-375 cell line was selected to be used in the following experiments.
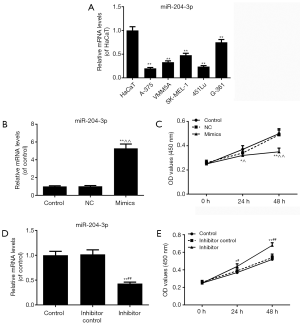
We transfected miR-204-3p mimic vectors into A-375cells to observe the changes of melanomas cell viability. As shown in Figure 1B, miR-204-3p level was observably higher than that in control group (P<0.01). The results of CCK-8 assay showed that the inhibitory effects of the overexpression of miR-204-3p on melanomas cell viability was increased over time, however, the viabilities were markedly reduced at 24 h (P<0.05) and 48 h (P<0.01) (Figure 1C). After the transfection of miR-204-3p inhibitor, the transfection sufficiency was shown in Figure 1D, and the miR-204-3p inhibition could significantly promote the cell viability of A-375 cells (P<0.01, Figure 1E).
MiR-204-3p expression negatively regulated the migration and invasion abilities of A-375 cells
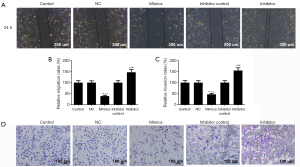
The wound healing and Transwell chamber assays were used to determine the effects of miR-204-3p on the migration and invasion abilities of malignant melanomas. The wound healing assay showed that the displaced distance of miR-204-3p mimic vector group was far below that of control group after 24 h (Figure 2A,B). Moreover, the number of invaded cells in miR-204-3p mimics group was markedly less than that in the control group (P<0.01, Figure 2C,D). Compared with the miR-204-3p overexpression, the transfection of miR-204-3p inhibitor, both the migration and invasion abilities of A375 cells were evidently enhanced (P<0.01). These results suggested that a negative correlation of miR-204-3p expression with the migration and invasion abilities of malignant melanomas.
MiR-204-3p could effectively target PAX2 gene
The results of target prediction by TargetScan and miRTarBase revealed that PAX2 and GNAQ were the target genes for the miR-204-3p, and we also found the position of 681-687 of PAX2-3’ UTR harbors one potential binding site for miR-204-3p (Figure 3A). We constructed a dual luciferase reporter plasmid to further verify the relation between miR-204-3p and PAX2 or GNAQ. As shown in Figure 3B, the luciferase activity of cells transfected with PAX2-3'UTR alone was higher than that in MiR-204-3p + PAX2-3'UTR group. After being co-transfected with GV272-miR-204-3p and GV268-PAX2-3'UTR plasmids, the luciferase activity was observably decreased. However, the luciferase activity had no difference from control + PAX2-3'UTR when GV268-PAX2-3'UTR was replaced by mutant 3'-UTR plasmid in the co-transfected group (Figure 3B). In addition, as Figure 3C shown, miR-204-3p hardly had any effects on the expression of GNAQ-3'UTR.
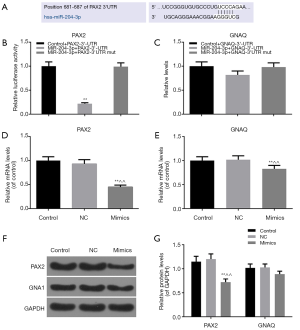
We also measured the transcriptional and posttranscriptional levels of PAX2 and GNAQ in the miR-204-3p mimics cells, and the results were consistent with dual luciferase report system (Figure 3D,E,F). Overexpression of miR-204-3p significantly inhibited the mRNA and protein levels of PAX2 (P<0.01, Figure 3D,E,F,G) and slightly reduced GNAQ mRNA level. Thus, these data suggested that miR-204-3p could directly modulated PAX2 through interaction with the binding site in its 3'-UTR.
Overexpression of miR-204-3p attenuated the expressions of EMT-associated genes in A-375 cells
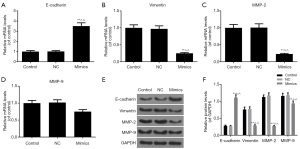
According to the inhibitory effects on miR-204-3p on the migration and invasion capacities of A-375 cells, we analyzed several EMT-associated genes expressions. As the results of qRT-PCR shown in Figure 4A,B,C,D, the E-cadherin mRNA level was notably elevated in the mimics group and the expressions of vimentin, MMP-2 and MMP-9 were notably downregulated (P<0.01). Furthermore, the result of Western blot was consistent with that of qRT-PCR (Figure 4E,F). Taken together, miR-204-3p overexpression had the ability of suppressing the EMT progression of malignant melanoma cells.
siPAX2 reversed the effects of miR-204-3p inhibition on A375 cell apoptosis
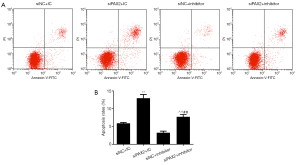
To confirm the involvement of PAX2 in miR-204-3p-induced regulatory effects on A375 cells, siPAX2 was introduced to co-transfect with miR-204-3p inhibitor, subsequently, detecting the expression of PAX2 and cell apoptosis. As shown in Figure 5A,B,C, miR-204-3p inhibition could notably up-regulate the mRNA and protein levels of PAX2. After co-transfection with siPAX2, the expression of PAX2 was observably decreased again (P<0.01). Moreover, we also detected the changes in cell apoptosis, and we found that miR-204 inhibition significantly inhibited the apoptosis, and the co-transfection of siPAX2 could notably reverse the effects of miR-204-3p inhibition (P<0.01, Figure 6A,B). Therefore, PAX2 played an important role in the regulatory effects of miR-204-3p on the A375 cells.

Discussion
Melanoma is the most dangerous type of skin cancer and seriously threatens the health of human life. It is estimated that an individual had a 2% lifetime risk of developing melanoma. As the increasing incidence occurred in recent years, malignant melanoma has attracted much research attention and generated considerable recent research interests (23). The leading cause of melanoma death is that melanoma cells invade neighboring tissue when the tissue is still markedly small in size just because of which let invasion capacity interference became one of the main directions in the treatment of melanoma. As reported previously, PAX2 was aberrantly expressed with strong ties to the cell survival and invasion of melanoma cells, which contributed to the EMT progression and ultimately induced tumor metastasis (24). In the current study, we found that the levels of miR-204-3p in several melanoma cell lines were generally lower than those in HaCaT cells. More importantly, we also verified that miR-204-3p could directly interact with the 3'UTR of PAX2 gene, thus, we measured some EMT-associated factors to further confirm the relation between miR-204-3p and EMT.
Growing researches indicated that miRNAs not only widely participated in cell proliferation, differentiation, development, apoptosis, inflammation and some other essential cellular processes (25), but also played crucial roles in tumor genesis and progression (26). In 2015, several miRNAs including miR-204, miR-141, miR-200 and miR-200c were found significantly decreased in melanoma and functioned as melanoma inhibitors (27). Meanwhile, miR-204 could target IGFBP5 and RAB22A and suppress the proliferation of melanoma cells to fulfill the inhibitory role (21,28). These researches were consistent with our results, as the viability of A-375 cells transfected with miR-204-3p mimic vectors were observably reduced, which may explain why the expression of miR-204-3p was markedly decreased after normal melanocytes having been transformed into malignant melanomas, further verifying that it had a tumor inhibitory role. To further explore how miR-204-3p acted on melanoma cells, we measured the migration and invasion capacities of melanoma cells after transfecting miR-204-3p overexpression vector, and the results suggested miR-204-3p had an effective inhibition on both abilities of malignant melanoma. In the research of hepatocellular carcinoma, miR-204-3p overexpression could inhibit the tumor endothelial cells growth by targeting fibronectin 1 (FN1), which was the essential glycoprotein macromolecule of extracellular matrix and played important roles in cellular adhesion, invasion and injury repair (29). In our results, we found that PAX2 was a potential target gene for miR-204-3p, however, the expression of PAX2 has been proved to be crucial for the tumor cells growth and invasion (24,30). Ueda et al. (31,32) demonstrated that PAX2 expression enhanced the invaded ability of prostate cancer cells via hepatocyte growth factor pathway. Similarly, overexpression of PAX2 also contributed to the metastasis ability of esophageal cancer (33). According to these results, we suggested that miR-204-3p overexpression markedly suppressed the cell viability, migration and invasion of malignant melanoma, and that regulating the expression of PAX2 might be a novel approach to inhibiting the metastatic melanoma.
Previous researches indicated that EMT was the prerequisite for the migration and invasion abilities of tumor cells (34,35). EMT is a complex biological process, and loss of E-cadherin is one of its major characteristics, which could result in decreased cell-cell adhesion and enhanced aggressiveness of epithelial cells. Vimentin is another marker of EMT, and the upregulation of vimentin and downregulated E-cadherin were closely associated with the migration and invasion of tumor cells (36,37). Previous study showed that MMP-2 and 9, the members of MMPs family, are the main enzyme to degrade extra cellular matrix (ECM) and promote cancer invasion and metastasis (38). Our results suggested that overexpression of miR-204-3p had a negative effect on the EMT progression of A-375 cells through increasing E-cadherin expression and attenuating the mRNA and protein levels of vimentin, MMP-2 and MMP-9. More interestingly, in 2017, Yang et al. (39) revealed that matrine could inhibit the invasion and metastasis of non-small-cell lung cancer (NSCLC) through decreasing PAX2 expression. Based on this conclusion and current results, we speculated that miR-204-3p overexpression inhibited the migration and invasion capacities and the EMT progression of melanoma cells through targeting PAX2.
However, there are some shortages remained in our study. For example, extending our experimental results to another type of melanoma cell line could validate our hypothesis better. Moreover, the clinical association analysis of miR-204-3p and PAX2 in melanoma can be able to reveal well the significance of our study.
Conclusions
In conclusion, our findings showed that miR-204-3p overexpression was negatively correlated with the migration and invasion abilities of melanoma cells. For the first time, we found that miR-204-3p could directly modulate the expression of PAX2 by binding its 3'UTR, and the knockdown of PAX2 had an inhibitory effect of miR-204-3p on the cell apoptosis malignant melanoma cells. Finally, our findings propose that miR-204-3p could destroy the bridge between PAX2 and migration and invasion ability of malignant melanoma, and suggest that miR-204-3p might be used as an effective target for the treatment of metastatic melanoma.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.10). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and individual informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014;64:9-29. [Crossref] [PubMed]
- Aftab MN, Dinger ME, Perera RJ. The role of microRNAs and long non-coding RNAs in the pathology, diagnosis, and management of melanoma. Arch Biochem Biophys 2014;563:60-70. [Crossref] [PubMed]
- Orgaz JL, Sanz-Moreno V. Emerging molecular targets in melanoma invasion and metastasis. Pigment Cell Melanoma Res 2013;26:39-57. [Crossref] [PubMed]
- Pessina F, Navarria P, Tomatis S, et al. Outcome Evaluation of Patients with Limited Brain Metastasis From Malignant Melanoma, Treated with Surgery, Radiation Therapy, and Targeted Therapy. World Neurosurg 2017;105:184-90. [Crossref] [PubMed]
- Seo Y, Baba H, Fukuda T, et al. High expression of vascular endothelial growth factor is associated with liver metastasis and a poor prognosis for patients with ductal pancreatic adenocarcinoma. Cancer 2000;88:2239-45. [Crossref] [PubMed]
- Seto S, Onodera H, Kaido T, et al. Tissue factor expression in human colorectal carcinoma: correlation with hepatic metastasis and impact on prognosis. Cancer 2000;88:295-301. [Crossref] [PubMed]
- Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science 2011;331:1559-64. [Crossref] [PubMed]
- Chiang AC, Massagué J. Molecular basis of metastasis. N Engl J Med 2008;359:2814-23. [Crossref] [PubMed]
- Rigel DS, Russak J, Friedman R. The evolution of melanoma diagnosis: 25 years beyond the ABCDs. CA Cancer J Clin 2010;60:301-16. [Crossref] [PubMed]
- Hendrix MJ, Seftor EA, Seftor RE, et al. Reprogramming metastatic tumour cells with embryonic microenvironments. Nat Rev Cancer 2007;7:246-55. [Crossref] [PubMed]
- Jiang H, Li L, Yang H, et al. Pax2 may play a role in kidney development by regulating the expression of TBX1. Mol Biol Rep 2014;41:7491-8. [Crossref] [PubMed]
- Ranghini EJ, Dressler GR. Evidence for intermediate mesoderm and kidney progenitor cell specification by Pax2 and PTIP dependent mechanisms. Dev Biol 2015;399:296-305. [Crossref] [PubMed]
- Kaur G, Li CG, Chantry A, et al. SMAD proteins directly suppress PAX2 transcription downstream of transforming growth factor-beta 1 (TGF-beta1) signalling in renal cell carcinoma. Oncotarget 2018;9:26852-67. [PubMed]
- Al-Hujaily EM, Tang Y, Yao DS, et al. Divergent Roles of PAX2 in the Etiology and Progression of Ovarian Cancer. Cancer Prev Res (Phila) 2015;8:1163-73. [Crossref] [PubMed]
- Wang M, Ma H, Pan Y, et al. PAX2 and PAX8 reliably distinguishes ovarian serous tumors from mucinous tumors. Appl Immunohistochem Mol Morphol 2015;23:280-7. [Crossref] [PubMed]
- Li L, Wu Y, Yang Y. Paired box 2 induces epithelial-mesenchymal transition in normal renal tubular epithelial cells of rats. Mol Med Rep 2013;7:1549-54. [Crossref] [PubMed]
- Tellez CS, Shen L, Estecio MR, et al. CpG island methylation profiling in human melanoma cell lines. Melanoma Res 2009;19:146-55. [Crossref] [PubMed]
- Thyagarajan A, Shaban A, Sahu RP. MicroRNA-Directed Cancer Therapies: Implications in Melanoma Intervention. J Pharmacol Exp Ther 2018;364:1-12. [Crossref] [PubMed]
- Xin Y, Li Z, Chan MT, et al. Circulating epigenetic biomarkers in melanoma. Tumour Biol 2016;37:1487-92. [Crossref] [PubMed]
- Xia Y, Zhu Y, Ma T, et al. miR-204 functions as a tumor suppressor by regulating SIX1 in NSCLC. FEBS Lett 2014;588:3703-12. [Crossref] [PubMed]
- Liu L, Wang J, Li X, et al. MiR-204-5p suppresses cell proliferation by inhibiting IGFBP5 in papillary thyroid carcinoma. Biochem Biophys Res Commun 2015;457:621-6. [Crossref] [PubMed]
- Tao K, Yang J, Guo Z, et al. Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am J Transl Res 2014;6:391-401. [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Lee SB, Doberstein K, Baumgarten P, et al. PAX2 regulates ADAM10 expression and mediates anchorage-independent cell growth of melanoma cells. PLoS One 2011;6:e22312. [Crossref] [PubMed]
- Brennecke J, Hipfner DR, Stark A, et al. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 2003;113:25-36. [Crossref] [PubMed]
- Min S, Liang X, Zhang M, et al. Multiple tumor-associated microRNAs modulate the survival and longevity of dendritic cells by targeting YWHAZ and Bcl2 signaling pathways. J Immunol 2013;190:2437-46. [Crossref] [PubMed]
- Li C, Wang X, Chen YN, et al. MicroRNA-21, 204 and 125b Play Potential Roles in Tumorigenesis of Melanoma. Advances in Bioscience & Biotechnology 2015;6:677-92. [Crossref]
- Yin Y, Zhang B, Wang W, et al. miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clin Cancer Res 2014;20:6187-99. [Crossref] [PubMed]
- Cui ZH, Shen SQ, Chen ZB, et al. Growth inhibition of hepatocellular carcinoma tumor endothelial cells by miR-204-3p and underlying mechanism. World J Gastroenterol 2014;20:5493-504. [Crossref] [PubMed]
- Busse A, Rietz A, Schwartz S, et al. An intron 9 containing splice variant of PAX2. J Transl Med 2009;7:36. [Crossref] [PubMed]
- Ueda T, Ito S, Shiraishi T, et al. Hyper-expression of PAX2 in human metastatic prostate tumors and its role as a cancer promoter in an in vitro invasion model. Prostate 2013;73:1403-12. [Crossref] [PubMed]
- Ueda T, Ito S, Shiraishi T, et al. PAX2 promoted prostate cancer cell invasion through transcriptional regulation of HGF in an in vitro model. Biochim Biophys Acta 2015;1852:2467-73. [Crossref] [PubMed]
- Liu P, Gao Y, Huan J, et al. Upregulation of PAX2 promotes the metastasis of esophageal cancer through interleukin-5. Cell Physiol Biochem 2015;35:740-54. [Crossref] [PubMed]
- Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res 2006;66:8319-26. [Crossref] [PubMed]
- Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med 2006;12:895-904. [Crossref] [PubMed]
- Qureshi R, Arora H, Rizvi MA. EMT in cervical cancer: its role in tumour progression and response to therapy. Cancer Lett 2015;356:321-31. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Itoh T, Tanioka M, Matsuda H, et al. Experimental metastasis is suppressed in MMP-9-deficient mice. Clin Exp Metastasis 1999;17:177-81. [Crossref] [PubMed]
- Yang J, He D, Peng Y, et al. Matrine suppresses the migration and invasion of NSCLC cells by inhibiting PAX2-induced epithelial-mesenchymal transition. Onco Targets Ther 2017;10:5209-17. [Crossref] [PubMed]


