
Silencing NEDD9 by lentivirus-delivered shRNA inhibits the growth of BxPC-3 cells in vitro and in vivo
Introduction
Pancreatic cancer (PC) is an aggressive malignant tumor of the digestive system that has high rates of invasion and early metastasis. Due to its rapid invasion and early metastasis, most PCs are resistant to current standard therapies (1). PC prognosis is very poor with a median survival time of fewer than 6 months and a 5-year survival rate below 6% (2). Thus, there is an urgent need to elucidate the underlying molecular mechanisms of PC initiation, progression, and maintenance to identify novel therapeutic strategies.
The neural precursor cell-expressed, developmentally downregulated protein 9 (NEDD9), also known as HEF1 and Cas-L, is a non-catalytic scaffolding protein and a member of the Crk-associated substrate (CAS) protein family (3-5). It plays an essential role in the regulation of cell proliferation, adhesion, apoptosis, differentiation, and invasion (6,7). It has been reported that NEDD9 is overexpressed in numerous types of cancer, including breast cancer, lung cancer, gastric cancer, melanoma, and glioblastoma (8-12), suggesting that the abnormal expression of NEDD9 is significantly related to the occurrence and development of cancers. Further studies have indicated that a decrease in the expression of NEDD9 through RNA interference could substantially suppress proliferation, migration, and invasion of melanoma, glioma, and gastric cancer cells (8,13,14).
In our previous study, we found that NEDD9 was overexpressed in PC tissues compared to adjacent noncancerous pancreatic tissues, and that a high expression level of NEDD9 was significantly correlated with clinical staging, lymph node metastasis, and histological differentiation (15). Patients with a higher NEDD9 expression had a significantly shorter survival time than those patients with a lower NEDD9 expression, suggesting that NEDD9 is a potential biomarker for the diagnosis and prognosis of PC, and may be a novel target for anticancer therapies. In the present study, in order to identify whether RNA interference (RNAi) could specifically suppress the expression of target genes and exert distinct biological action in PC tumorigenesis, we investigated NEDD9 expression and its functional roles in human PC BxPC-3 cells in vitro and in vivo following RNAi-mediated silencing of NEDD9.
Methods
Cell lines selection
Five kinds of cell lines (CFPAC-1, PANC-1, BxPC-3, ASPC-1, and Pacn-02) were obtained from the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPIM1640 medium (Gibco, CA, USA) supplemented with 10% fetal bovine serum (FBS) (HyClone, LA, USA) at 37 °C in a humidified atmosphere containing 5% CO2. Quantitative RT-PCR and western blot were used to detect the expression of NEDD9 mRNA and protein levels in the above cell lines. The cell line which expressed NEDD9 the most was selected to carry out the following experiment.
NEDD9 RNAi plasmid construct
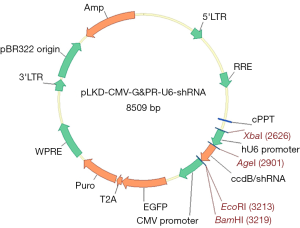
To determine the most effective interference target, 3 interference targets were designed and selected. The BxPC-3 cells were divided into 5 groups: blank control (normal BxPC-3 cells), negative control (Y007), NEDD9-shRNA1 (Y2600), NEDD9-shRNA2 (Y2601), and NEDD9-shRNA3 (Y2602). The oligonucleotides for the short hairpin (sh)RNA targeting the sequence of NEDD9 (Y007: TTCTCCGAACGTGTCACGT; Y2600: GCTCTCAGAACGACGCATATG; Y2601: GCAGCTGGTCCCTGAATATCT; Y2602: GCAGGAAATGGTGCACCAAGT; NCBI Gene ID: 4739) were chemically synthesized by Obio Technology (Shanghai, China) and cloned into the vector pLKD-CMV-G&PR-U6-shRNA (Obio Technology, Shanghai, China) to get plasmid pLKD-CMV-G&PR-U6-NEDD9-shRNA (Figure 1). The sequence of the plasmid was confirmed by direct DNA sequencing.
Establishment of stable NEDD9 shRNA BxPC-3 cells
BxPC-3 cells were plated in 24-well plates (2×104 cells/well) in RPIM1640 (Gibco, CA, USA) supplemented with 10% FBS (HyClone, LA, USA). Every well contained 5 µg/mL polybrene (HyClone, LA, USA) and was incubated at 37 °C in 5% CO2. The pLKD-CMV-G&PR-U6-NEDD9 shRNA together with the packaging plasmids were co-transfected into 293T cells to obtain a retrovirus. The retrovirus was transduced into BxPC-3 cells and then selected with puromycin to obtain stable NEDD9-shRNA-expressing BxPC-3 cells. The same procedures obtained control BxPC-3 cells stably transduced with pLKD-CMV-G&PR-U6-shRNA. Green fluorescent protein (GFP) expression was observed by fluorescence microscopy 14 d after transduction, and cells were harvested at 14 d after transduction for western blot analysis.
Western blot
Cells were lysed in NP-40 lysis buffer (Beyotime, Shanghai, China) on ice for 20 min. Protein concentrations were determined using a BCA protein assay kit (HyClone-Pierce, LA, USA). Protein samples (36 µg) were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto membranes (Millipore, USA). The membranes were blocked with 5% non-fat milk in Tris-buffered saline and Tween 20 (10 mM Tris-HCl, pH 8.0, 100 mM NaCl and 0.05% Tween, TBS-T). The membranes were then probed with mouse anti-NEDD9 antibody (1:2,000) (Abcam, MA, USA) or anti-GAPDH antibody (1:10,000) (Beyotime, Shanghai, China) at 4 °C overnight. Membranes were washed 3 times with TBS-T and incubated with horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:3,000) (Santa Cruz Biotechnology, USA) for 2 h at room temperature. The immunohistochemical reaction of bound antibodies was visualized using ECL PlusTM western blotting system (Amersham Pharmacia Biotech, UK).
Quantitative RT-PCR
Total RNA from BxPC-3cells was extracted using TRIzol reagent (Invitrogen, CA, USA) according to the manufacturer’s instructions. cDNA was synthesized with M-MLV retroviridase (Promega, WI, USA). Primers were synthesized by Obio Technology (Shanghai, China). The sequences of the used primers were as follows: NEDD9, upstream 5'-TTACGTCCACCTACAGGGTA-3', downstream 5'-CGGGCTTTGTAATCTCTTG-3'; β-actin, upstream 5'-TTCTACAATGAGCTGCGTG-3', downstream 5'-CTCAAACATGATCTGGGTC-3'. qRT-PCR was performed on the ABI PRISM 7500 Sequence Detection System (Biosystems, CA, USA). β-actin was used as an endogenous control. PCR reactions were performed in triplicate, and data were analyzed through the comparative threshold cycle (CT) method.
Colony formation assay
Stably transfected cells were cultured in RPIM1640 (Gibco, CA, USA) supplemented with 10% FBS (HyClone, LA, USA) in 6-well plates. The cells were plated at an initial density of 1,000 cells/well. The medium was replaced with 10 mL fresh medium every 3 d. After 14 d of culture, the cells were fixed with methanol and stained with 0.5% crystal violet (Beyotime, Shanghai, China) for 30 min. Colonies with more than 50 cells were counted, and photos were taken using inverted fluorescence microscopy.
Cell migration and invasion assays
The migration and invasion of BxPC-3 cells were quantified by wound healing and Transwell assays, respectively. For wound healing assay, cells (5×105 cells/well) were plated in 6-well plates. After cells became 80–90% confluent, the “wound” was scratched manually with a 200-µL pipette tip. Images were taken at ×100 magnification fields from each 6-well plate at 0, 16, and 24 h. For Transwell assay, cells were placed in the upper chamber of Transwells (8 µmol/L pore size polycarbonate membranes) (Corning, CA, USA) with serum-free RPIM1640 medium. Lower chambers were loaded with 500 µL RPIM1640 containing 10% FBS. After 24 h of incubation at 37 °C in 5% CO2, non-migratory cells were removed. Invasive cells were fixed with 4% paraformaldehyde for 15 min and stained with 500 µL crystal violet. After 3 washes with ddH2O, 3 fields per well were photographed under phase-contrast microscopy. The number of cells per field was counted.
In vivo study of pancreatic xenograft tumor models in nude mice
Animal experiments were approved by the Third Affiliated Hospital of Nantong University Medical Ethics Committee. Six-week-old male BALB/c nude mice (N=18) were purchased from Shanghai SLAC Laboratory Animal (Shanghai, China). The mice were randomly divided into 3 groups (n=6 per group): BxPC-3 control (blank), negative-shRNA (negative), and NEDD9-shRNA (shRNA) groups. Cells (1×107 in 0.2 mL RPIM1640 supplemented with 10% FBS) were injected into the right flank of the mice. The tumor volume was measured every 3–4 d. Tumor volume was calculated using the formula: width2 (mm2) × length (mm)/2 every 3–4 d using a caliper. After 5 weeks, the mice were sacrificed, and tumors were harvested and weighed.
Statistical analysis
Each experiment was conducted at least 3 times. Values are presented as mean ± standard deviation (SD). Statistical analysis of the data was performed using a Student’s t-test with IBM SPSS Statistic v19.0 (IBM Co., Armonk, NY, USA). P<0.05 indicates a significant difference, and P<0.01 indicates a highly significant difference.
Results
In vitro study
Cell line selection
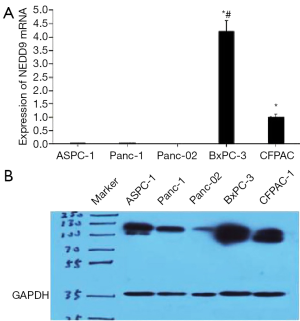
Five kinds of pancreatic tumor cells (CFPAC-1, PANC-1, BxPC-3, ASPC-1 and Panc-02 cell lines) were selected to detect the expression of NEDD9. According to the result of qPCR, the BxPC-3 cells expressed NEDD9 the most while no expression was detected with Panc-02 (Figure 2A). From the western blot result, 2 phosphorylated isoforms of NEDD9, migrating at 105 kDa (p105) and 115 kDa (p115), were detected (Figure 2B) while NEDD9 was expressed the most in the BxPC-3 cells which conformed with the qPCR result. Consequently, BxPC-3 cells were selected to carry out the following experiment.
Establishment of stable BxPC-3 cells expressing NEDD9 shRNA
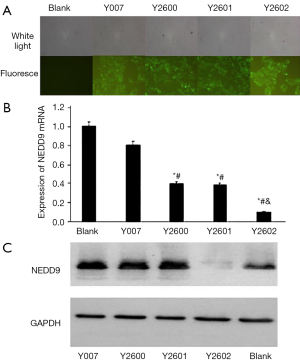
Four shRNA sequences (Y007, Y2600, Y2601, and Y2602) in plasmid pLKD-CMV-G&PR-U6-NEDD9-shRNA and the cloning sites in vector pLKD-CMV-G&PR-U6-shRNA were confirmed by DNA sequencing (Figure 3). The plasmids were transfected into 293T cells to pack into lentivirus NEDD9-RNAi and negative control shRNA, respectively. The titer of viruses was 3.04×108 titer U/mL. After NEDD9-shRNA and nonspecific shRNA, marked by fluorescence, were transfected into BxPC-3 cells, the cells were selected with puromycin for 14 d to obtain stable BxPC-3 cells expressing NEDD9 shRNA. Cells showing green fluorescence were considered successfully transduced. Significant GFP was observed in the shRNA transfection group and the negative control group while there was no fluorescence in the blank control group. This indicated that NEDD9 shRNA had been successfully transfected into BxPC-3 cells (Figure 4A). The stably transduced cell lines were confirmed by qRT-PCR and western blot for detecting the expression of NEDD9 mRNA and protein levels, respectively. We found that NEDD9-Y2602 shRNA inhibited the expression of NEDD9 significantly (Figure 4B,C); therefore, it was selected as the best shRNA with which to carry out the interference experiment.
Silencing of NEDD9 inhibits proliferation, migration, and invasion of BxPC-3 cells
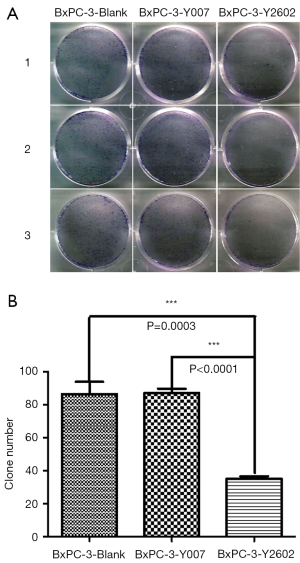
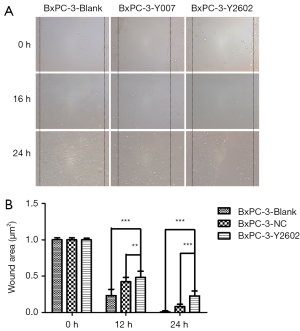
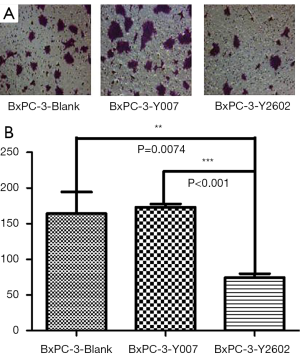
To further characterize the role of NEDD9-shRNA in BxPC-3 cells, we performed colony formation assays to assess the effect of NEDD9-shRNA on the proliferation of BxPC-3 cells. After culturing for 14 d, the NEDD9-RNAi group showed significantly fewer clones compared to the blank and negative control groups (P<0.001). No significant difference was found between the blank and negative control group (P>0.05) (Figure 5A,B). Moreover, the wound-healing assay showed that cell migration was significantly decreased in the RNAi group compared to the blank and negative control groups (P<0.05; Figure 6A,B). The Transwell assay demonstrated that there were significantly fewer invasive cells in the NEDD9-RNAi group compared to the blank and negative control groups (Figure 7A,B, P<0.01). These results demonstrate that silencing NEDD9 by RNAi reduced the proliferation, migration, and invasion of BxPC-3 cells.
In vivo study
NEDD9-RNAi suppresses the growth of xenograft tumor in nude mice
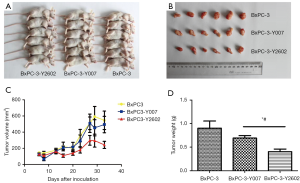
To determine whether knockdown of NEDD9 inhibits pancreatic tumor growth in vivo, BxPC-3 control (blank), negative-shRNA (negative), and NEDD9-shRNA (shRNA) cells were injected into the right flank of the BALB/c nude mice. The growth of pancreatic adenocarcinoma tumor xenografts in the 3 treatment groups was compared. Measurement of tumor volumes began once subcutaneous tumors became palpable and continued until day 35, at which point tumors were harvested and weighed (Figure 8A,B). The average volume of tumors in the NEDD9-shRNA group was significantly smaller compared to the volume of tumors in the16 negative or blank groups (P<0.05; Figure 8C). In addition, xenografts in the NEDD9-RNAi group had markedly lower tumor weights (0.4±0.13 g) on day 35 compared to xenografts of the negative (0.7±0.12 g) (P<0.05) or blank (0.9±0.37 g) (P<0.05) groups (Figure 8D). Thus, silencing the expression of NEDD9 by RNAi can inhibit the growth of the BxPC-3 xenografts in nude mice.
Discussion
PC is the 12th most common malignancy and the 7th leading cause of cancer mortality, accounting for more than 330,000 deaths in 2012 (16). Recent studies have suggested that the incidence and mortality of PC are rising in many countries, and some projection studies have estimated that PC will escalate from the 4th to the 2nd leading cause of cancer death in the USA by 2020 (17). It has already induced a substantial globe burden which needs to be taken seriously (18,19). Owing to its extremely aggressive nature and reduced survival rate (20), the underlying molecular mechanisms of PC urgently need to be clarified in order to innovate novel therapeutic strategies.
NEDD9, a skeletal protein belonging to the Crk-associated substrate (CAS) family, acts as a router in the cell signal transduction process and plays a vital role in the regulation of cell cycle-related events such as DNA replication, chromatin condensing, and segregation (21,22). Accumulating evidence has shown that NEDD9 plays an essential role in multiple steps of tumorigenesis for various cancers, and that deregulation of NEDD9 is closely associated with the pathophysiology of several types of cancers, including breast, lung, and gastric cancers. For instance, NEDD9 overexpression promotes migration and invasion of breast tumor cells through multiple mechanisms including activating the focal adhesion complex (FAK and SRC), mediating effects of TGFβ and integrins, and increasing synthesis of tumor-associated glycocalyx (9,23,24). In lung and gastric cancers, elevated expression of NEDD9 have been closely associated with malignant progression and metastases (10,11). In our previous study, we investigated the expression of NEDD9 in 106 cases of PC and paired adjacent healthy tissues by RT-PCR, western blotting, and immunohistochemistry. We found that NEDD9 was upregulated in PC tissues, but weakly expressed in adjacent healthy tissues.
Moreover, the clinicopathological analysis revealed that NEDD9 protein expression was significantly correlated with TNM stage, tumor differentiation, and metastasis (15). Taken together, this evidence suggests that NEDD9 may act as an oncogene and participates in the invasion and metastasis in PC. We, therefore, studied the effect of inhibiting expression of NEDD9 on pancreatic adenocarcinoma BxPC-3 cells both in vivo and in vitro.
However, NEDD9 is a scaffold protein with no apparent enzymatic functions, which poses a challenge for developing small molecules that can inhibit the functions of NEDD9. RNA interference technology is a new silencing technology which involves the insertion into cells of endogenous or exogenous double-stranded RNA. It can efficiently suppress the expression of genes and proteins (25). As a method of RNAi, endogenously expressed shRNAs have been completed through various viral systems, including retrovirus, adenovirus, adeno-associated virus (AAV), and lentivirus (26). The viral systems have a clear advantage of high transduction efficiency and stable expression of shRNAs for a prolonged period. So, in this study, we developed a lentivirus-delivered shRNA system to efficiently reduce the expression of NEDD9 in a pancreatic adenocarcinoma BxPC-3 cell model. We demonstrated that lentivirus-delivered shRNA reduced NEDD9 expression in pancreatic adenocarcinoma BxPC-3 cells efficiently and stably for an extended period. This suggests that lentivirus-delivered shRNA may be a potential agent for inhibiting targeted genes.
In this study, we selected 3 shRNA sequences of siRNA which was targeting NEDD9 and BxPC-3 cells by lentivirus to silence NEDD9 in cells. Expression of mRNA was detected by qTR-PCR while protein was detected by western blot. The results showed that NEDD9-shRNA Y2602 elicited the maximum decrease among the 3 cell lines; therefore, we selected NEDD9-shRNA Y2602 for further experiment.
We also examined cell proliferation, invasion, and migration using cell colony formation, scratch wound healing, and Transwell assays after silencing NEDD9 by shRNA. The results showed that NEDD9-shRNA Y2602 inhibited proliferation, migration, and invasion of BxPC-3 cells in vitro. More importantly, we injected (shRNA) cells into the right flank of BALB/c nude mice to determine whether knockdown of NEDD9 inhibits pancreatic tumor growth in vivo. We found that silencing NEDD9 by RNAi significantly reduced the growth of BxPC-3 cells in mouse xenografts as well. Given these findings, we propose NEDD9 maybe a direct target in PC treatment.
In summary, NEDD9 plays a vital role in the carcinogenesis of PC. In our study, inhibiting the expression of NEDD9 mRNA and protein by lentivirus-delivered shRNA could not only inhibit proliferation, migration, and invasion of pancreatic adenocarcinoma BxPC-3 cells in vitro, but also suppressed the growth of BxPC-3 cells in mouse xenografts. This suggests that NEDD9 may serve as a potential marker, and may act as an effective gene therapy of PC in the future. However, the specific mechanisms underlying the protective effects of NEDD9 RNAi against PC remain unknown, and further experiments should be carried out to investigate the therapeutic role of NEDD9 RNAi in treating PC.
Acknowledgments
Funding: Our research was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Animal experiments were approved by the Third Affiliated Hospital of Nantong University Medical Ethics Committee.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Muniraj T, Jamidar PA, Aslanian HR. Pancreatic cancer: a comprehensive review and update. Dis Mon 2013;59:368-402. [Crossref] [PubMed]
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014;64:9-29. [Crossref] [PubMed]
- Kumar S, Tomooka Y, Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem Biophys Res Commun 1992;185:1155-61. [Crossref] [PubMed]
- Law SF, Estojak J, Wang B, et al. Human enhancer of filamentation 1, a novel p130cas-like docking protein, associates with focal adhesion kinase and induces pseudohyphal growth in Saccharomyces cerevisiae. Mol Cell Biol 1996;16:3327-37. [Crossref] [PubMed]
- Minegishi M. Structure and function of Cas-L, a 105-kD Crk-associated substrate- related protein that is involved in beta 1 integrin-mediated signaling in lymphocytes. J Exp Med 1996;184:1365-75. [Crossref] [PubMed]
- Nikonova AS, Gaponova AV, Kudinov AE, et al. CAS proteins in health and disease: An update. IUBMB Life 2014;66:387-95. [Crossref] [PubMed]
- Singh MK, Cowell L, Seo S, et al. Molecular basis for HEF1/NEDD9/Cas-L action as a multifunctional co-ordinator of invasion, apoptosis and cell cycle. Cell Biochem Biophys 2007;48:54-72. [Crossref] [PubMed]
- Natarajan M, Stewart JE, Golemis EA, et al. HEF1 is a necessary and specific downstream effector of FAK that promotes the migration of glioblastoma cells. Oncogene 2006;25:1721-32. [Crossref] [PubMed]
- Kong C, Wang C, Wang L, et al. NEDD9 Is a Positive Regulator of Epithelial-Mesenchymal Transition and Promotes Invasion in Aggressive Breast Cancer. PLoS One 2011;6:e22666. [Crossref] [PubMed]
- Chang JX, Gao F, Zhao GQ, et al. Role of NEDD9 in invasion and metastasis of lung adenocarcinoma. Exp Ther Med 2012;4:795-800. [Crossref] [PubMed]
- Zhang SS, Wu LH, Liu Q, et al. Elevated expression of NEDD9 is associated with metastatic activity in gastric cancer. Onco Targets Ther 2015;8:633-40. [Crossref] [PubMed]
- Liu W, Monahan K, Pfefferle A, et al. LKB1/STK11 inactivation leads to expansion of a prometastatic tumor subpopulation in melanoma. Cancer Cell 2012;21:751-64. [Crossref] [PubMed]
- Zhang S, Wu L, Liu Q, et al. Impact on growth and invasion of gastric cancer cell lines by silencing NEDD9. Onco Targets Ther 2015;8:223-31. [Crossref] [PubMed]
- Kim M, Gans JD, Nogueira C, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell 2006;125:1269-81. [Crossref] [PubMed]
- Xue YZ, Sheng YY, Liu ZL, et al. Expression of NEDD9 in pancreatic ductal adenocarcinoma and its clinical significance. Tumour Biol 2013;34:895-9. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Ferlay J, Soerjomataram I, Ervik M, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase. No. 11. Lyon, France: International Agency for Research on Cancer, 2013.
- Wong MCS, Jiang JY, Liang M, et al. Global temporal patterns of pancreatic cancer and association with socioeconomic development. Sci Rep 2017;7:3165. [Crossref] [PubMed]
- Tseng CM, Huang SP, Liao WC, et al. Incidence and mortality of pancreatic cancer on a rapid rise in Taiwan, 1999-2012. Cancer Epidemiol 2017;49:75-84. [Crossref] [PubMed]
- Maisonneuve P, Lowenfels AB. Epidemiology of pancreatic cancer: an update. Dig Dis 2010;28:645-56. [Crossref] [PubMed]
- Bradshaw LN, Zhong J, Bradbury P, et al. Estradiol stabilizes the 105-kDa phospho-form of the adhesion docking protein NEDD9 and suppresses NEDD9-dependent cell spreading in breast cancer cells. Biochim Biophys Acta 2011;1813:340-5. [Crossref] [PubMed]
- Zheng M, McKeown-Longo PJ. Cell adhesion regulates Ser/Thr phosphorylation and proteasomal degradation of HEF1/NEDD9/CAS-L. J Cell Sci 2006;119:96-103. [Crossref] [PubMed]
- Bruna A, Greenwood W, Le Quesne J, et al. TGFβ induces the formation of tumour-initiating cells in claudinlow breast cancer. Nat Commun 2012;3:1055. [Crossref] [PubMed]
- Kozyulina PY, Loskutov YV, Kozyreva VK, et al. Prometastatic NEDD9 regulates individual cell Migration via caveolin-1-dependent 3trafficking of integrins. Mol Cancer Res 2015;13:423-38. [Crossref] [PubMed]
- Hammond SM, Bernstein E, Beach D, et al. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000;404:293-6. [Crossref] [PubMed]
- Liu YP, Berkhout B. miRNA cassettes in viral vectors: problems and solutions. Biochim Biophys Acta 2011;1809:732-45. [Crossref] [PubMed]


