
DNA hypermethylation of MAL gene may act as an independent predictor of favorable prognosis in patients with colorectal cancer
Introduction
Colorectal cancer (CRC) had a high estimated death of 881,000 in 2018. And the mortality ranked second in the world (1). The global burden of CRC is expected to reach 1,100,000 cancer deaths by 2030 (2). The 5-year relative survival rate for CRC patients has remained less than 50% in low-income countries (3). Surgery to remove the primary tumor is the most common treatment for CRC patients; however, approximately one-half of postoperative patients will experience a recurrence in the first three years (4). Survival of postoperative CRC patients is not only tightly related to pathological staging and specific histological features, but also related to multiple genetic and epigenetic alterations.
CRC is biologically a multi-step carcinogenic disease that is characterized by genetic and epigenetic disorders of signal transduction cascades. Various epigenetic mechanisms are involved in the tissue-specific expression and maintenance of chromosome stability, which provide opportunities for early detection, diagnosis, prognosis and management of CRC patients (5,6). DNA methylation, as the most popular epigenetic alteration, can regulate gene expression by the mechanisms which include chromatin complex alterations and recruitment of methyl-CpG domain-binding proteins around CpG islands (6). Large and sufficiently powered clinical studies have demonstrated the feasibility of using specific methylated DNA signatures as prognostic biomarkers in CRC tissues. The DNA hypermethylation of CDKN2A, EVL and IGFBP3 was associated with poor prognosis (7-9). Moreover, DNA methylation can reflect the expression of specific gene, which can be utilized as a surrogate for RNA- or protein-based expression analysis of tumor (10).
MAL gene encodes the integral membrane protein, which is a component of the protein machinery for raft remodeling by playing an essential role in T-cell development (11,12). MAL gene also encodes part of the protein machinery for glycolipid-enriched membrane-mediated apical transport in epithelial polarized cells, which was also important in membrane signal transduction (13-15). Although the expression of MAL was frequently decreased in gastric, esophageal and cervical carcinoma cells, it was increased by treatment with the demethylation reagent 5-aza-2'-deoxycytidine (16-18). MAL over-expression could decrease cellular motility and increase levels of apoptosis, which exhibited diminished tumorigenicity via the Fas signaling pathway (17). In addition, Zanotti et al. revealed that higher MAL mRNA levels were significantly associated with shorter overall survival (OS) and poor progression free survival (PFS) in high-grade serous ovarian carcinoma patients (19). Buffart et al. indicated that MAL hypermethylation in gastric cancer patients had a better survival compared with unmethylation patients (20). MAL hypermethylation was unequally distributed in a region close to the transcription start point and significantly associated with reduced gene expression in vitro in CRC (21,22). Therefore, we speculated that MAL methylation might act as a biomarker for prognosis of CRC patients. In the present study, we aimed to evaluate the CpG island methylation of MAL in CRC, and investigate the association between MAL methylation and prognosis of postoperative patients with CRC.
Methods
Patient cohort and materials collection
Two hundred and ninety-eight sporadic non-metastatic CRC patients (295 are Han, 2 are Manchu and 1 is Korean), confirmed by pathological diagnosis, were included in this study. The subjects were from a follow-up study of 453 patients who underwent surgical resection in the Third Affiliated Hospital of Harbin Medical University (Harbin, Heilongjiang, China) from November 2004 to July 2005 and from May 2007 to January 2008. Patients with any other cancer history were excluded. No patient had received pre-operative radiotherapy or chemotherapy before. The tumor tissues and adjacent non-tumor tissues (>10 cm distance to primary tumor) were immediately frozen in liquid nitrogen after resection and stored at –80 °C for further experiments. Clinical data were also collected, which included age at diagnosis, gender, tumor markers [preoperative carcinoembryonic antigen (CEA), preoperative CA19-9], clinicopathologic characteristics [tumor location, tumor-node-metastasis (TNM) stages, tumor invasion, lymph nodes involved, metastasis status, pathological classification, histological classification and differentiation degree] and clinical information of disease diagnosis and treatment (cancer detection methods, operation methods, the use of intestinal stapler, intraoperative chemotherapy, postoperative chemotherapy and postoperative radiotherapy). All patients provided informed consent at the time of sample collection. The study was approved by the Research Ethics Committee of Harbin Medical University.
The OS time was used as a measure of prognosis, which was defined as the time from surgery to the patient’s death of all causes or the last follow-up visit. The date of the last follow-up was March 15, 2014 (109 months).
DNA extraction and sodium bisulfate modification
Genomic DNA was extracted from tumor tissue and adjacent non-tumor tissue specimens using the classical phenol-chloroform procedure and stored at –80 °C.
Genomic DNA was bisulfate treated using a commercially available DNA modification kit (EpiTect Bisulfite Kit®, Qiagen, Hilden, Germany) and stored at –20 °C. All procedures were followed according to manufacturer’s instructions.
Methylation analysis of MAL
Methylation-sensitive high resolution melting (MS-HRM) was performed on a LightCycler 480 (Roche Applied Science, Mannheim, Germany) equipped with Gene Scanning software (version 2.0) to detect and analyze MAL methylation, as previously published (23). Universal unmethylated (0% methylated) and methylated (100% methylated) human whole genomic DNA samples (Zymo Research Corp., Irvine, CA, USA) were used as the unmethylated control and methylated DNA for MS-HRM analysis, respectively. A series of standards, including 100%, 50%, 20%, 10%, 5% and 0% methylated DNA (Figure 1), in a background of universal unmethylated DNA, were constructed by serially diluting the methylated control DNA into the unmethylated control according to mass concentration.
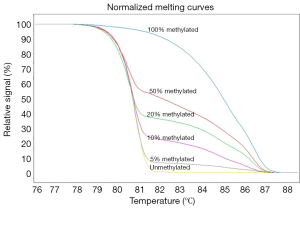
The MAL primers for MS-HRM analysis were as follows: forward primer, 5'-TGG TGA AGA TAG AGA AGT TAT TGG GTA GG-3'; and reverse primer, 5'-AAA ACC CCC AAA CCA CTA AAC TC-3', as reported previously (24). The amplification length was 160 bp, which was located at the CpG island close to the transcription start point of MAL gene. The whole reaction volume was 5.0 µL, which contained 2.5 µL of 1× LightCycler 480 High Resolution Melting Master Mix (Roche, Mannheim, Germany), 0.7 µL MgCl2 (25 mM), 0.1 µL of each forward and reverse primer(10 µM), 1.1 µL of polymerase chain reaction (PCR)-grade water and 0.5 µL of bisulfate-treated DNA. The cycling protocol started with one cycle at 95 °C for 10 min, followed by 50 cycles at 95 °C for 10 s, a touchdown of 64–58 °C for 30 s (0.4 °C/step) followed by 5 delays, 72 °C for 20 s, and a HRM step at 95 °C for 1 min, 40 °C for 1 min, and 70 °C for 5 s. The melting step was performed using a continuous acquisition between 70 and 93 °C at 40 acquisitions per 1 °C. Each plate included a water-blank as a negative control. A series of standard substances with known methylation ratios were included in each assay. To ensure the veracity and repeatability of the experiment, all samples were conducted independently in duplicate for each experiment. MS-HRM data were analyzed using Gene Scanning Software (Roche, Mannheim, Germany). Data processing included normalization and temperature shifting using LightCycler 480. The MAL methylation of tissue from CRC patients was determined by comparing curves of each sample to be tested with the series of standard substances in gene scanning module.
Statistical analysis
The cut-off value of MAL methylation was determined by using receiver operator characteristic (ROC) curve for permitting the clearest discrimination between tumor tissues and adjacent non-tumor tissues and for distinguishing hypermethylation from hypomethylation of tumor tissues. Missing values of the investigation data were generated using multiple imputation method provided by SPSS. All results were derived from pooled data of dataset obtained by multiple imputation method. The association between MAL methylation and clinicopathologic characteristics was evaluated with χ2 test. The survival rates were estimated using life table method. The comparison of survival rates between hypomethylation and hypermethylation groups was performed using log-rank tests. Associations of MAL methylation, clinicopathologic characteristics and clinical information of disease diagnosis and treatment with OS were analyzed by univariate Cox regression. Multivariate Cox regression was then employed to estimate the independent prognostic effect of MAL methylation. Hazard ratio (HR) and corresponding 95% confidence interval (CI) were calculated by Cox regression. Survival curve was constructed using the GraphPad Prism 7.0. All statistical analyses were performed using IBM SPSS Statistics 23.0 and two-sided P values <0.05 were considered to be statistically significant.
Propensity score (PS) method was used for accounting for baseline differences in characteristics between methylation groups. Multivariate logistic regression model was developed to estimate the PS, including the variables that are related to both outcomes and group decision or outcomes only.
Results
MAL methylation in tumor and adjacent non-tumor tissues
We examined MAL methylation of 298 primary tumor tissue specimens and 193 adjacent non-tumor tissue specimens. In tumor and adjacent non-tumor tissues, the median of MAL methylation were 20.0% (ranged from 0.0% to 60.0%) and 2.5% (ranged from 0.0% to 20.0%), respectively. The MAL methylation level of adjacent non-tumor tissues was significantly lower than tumor tissues (Mann-Whitney U test, P<0.001).
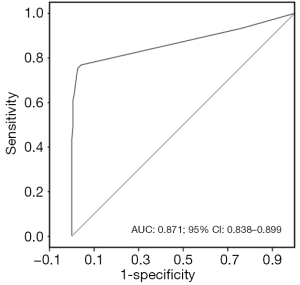
The cut-off value of MAL methylation was 2.5%, which had a high predictive ability to distinguish tumor tissues from adjacent non-tumor tissues. The sensitivity and specificity were 0.768 and 0.960, respectively, with the area under the curve (AUC) 0.871 (95% CI: 0.838–0.899) (Figure 2). There were 229 (76.8%) samples with MAL methylation >2.5% in tumor tissues, whereas there were only 8 (4.0%) samples in adjacent non-tumor tissues.
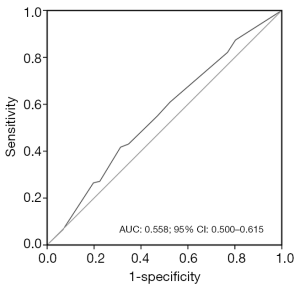
For further survival analysis, the cut-off value of MAL methylation for distinguishing the survival status was ≤7.5%, which meant that the tumor tissue with methylation ≤7.5% was defined as hypomethylation, others were defined as hypermethylation. The sensitivity and specificity were 0.422 and 0.689, respectively, with AUC 0.558 (95% CI: 0.500–0.615) (Figure 3). Thirty-six point six percent (109/298) CRC patients were in hypomethylation group, whereas 63.4% (189/298) CRC patients were in hypermethylation group.
The association between MAL methylation of tumor tissues and clinicopathologic characteristics of CRC patients
The median age of diagnosis for 298 CRC patients was 58 years old (ranging from 25–80 years old), and the male-to-female ratio was 1.42. The result showed that MAL methylation had no significant correlation with clinicopathologic characteristics (P>0.05) (Table 1).
Table 1
| Characteristics | Total (%) | MAL methylation, n (%) | P | |
|---|---|---|---|---|
| Hypomethylation (N=109) | Hypermethylation (N=189) | |||
| Age at diagnosis | 0.103 | |||
| <60 years | 162 (54.4) | 66 (60.6) | 96 (50.8) | |
| ≥60 years | 136 (45.6) | 43 (39.4) | 93 (49.2) | |
| Gender | 0.623 | |||
| Male | 175 (58.7) | 62 (56.9) | 113 (59.8) | |
| Female | 123 (41.3) | 47 (43.1) | 76 (40.2) | |
| CEA | 0.627 | |||
| <5 ng/mL | 123 (41.3) | 43 (39.4) | 80 (42.3) | |
| ≥5 ng/mL | 175 (58.7) | 66 (60.6) | 109 (57.7) | |
| CA19-9 | 0.711 | |||
| <37 U/mL | 217 (72.8) | 78 (71.6) | 139 (73.5) | |
| ≥37 U/mL | 81 (27.2) | 31 (28.4) | 50 (26.5) | |
| Tumor location | 0.953 | |||
| Colon | 110 (36.9) | 40 (36.7) | 70 (37.0) | |
| Rectum | 188 (63.1) | 69 (63.3) | 119 (63.0) | |
| TNM stages | 0.198 | |||
| I–II | 173 (58.1) | 58 (53.2) | 115 (60.8) | |
| III–IV | 125 (41.9) | 51 (46.8) | 74 (39.2) | |
| Tumor invasion | 0.279 | |||
| T1–T3 | 149 (50.0) | 50 (45.9) | 99 (52.4) | |
| T4 | 149 (50.0) | 59 (54.1) | 90 (47.6) | |
| Lymph nodes involved | 0.232 | |||
| N0 | 172 (57.7) | 58 (53.2) | 114 (60.3) | |
| N1–N2 | 126 (42.3) | 51 (46.8) | 75 (39.7) | |
| Metastasis status | 0.617 | |||
| M0 | 284 (95.3) | 103 (94.5) | 181 (95.8) | |
| M1 | 14 (4.7) | 6 (5.5) | 8 (4.2) | |
| Pathological classification | 0.157 | |||
| Prominence | 201 (67.4) | 68 (62.4) | 133 (70.4) | |
| Others | 97 (32.6) | 41 (37.6) | 56 (29.6) | |
| Histologic classification | 0.841 | |||
| Adenocarcinoma | 227 (76.2) | 85 (78.0) | 142 (75.1) | |
| Mucinous adenocarcinoma | 60 (20.1) | 20 (18.3) | 40 (21.2) | |
| Others | 11 (3.7) | 4 (3.7) | 7 (3.7) | |
| Differentiation degree | 0.424 | |||
| Poor | 48 (16.1) | 20 (18.3) | 28 (14.8) | |
| Moderate or well | 250 (83.9) | 89 (81.7) | 161 (85.2) | |
CEA, carcinoembryonic antigen; TNM, tumor-node-metastasis.
MAL methylation in tumor tissues and prognosis of CRC patients
At the end of the 109 months’ follow-up, 41.3% (123/298) of the patients died and 45.0% (134/298) were still alive. The median OS time was 58 months for hypomethylation group, 74 months for hypermethylation group and 73 months for all patients (the follow-up time ranged from 1–109 months). In hypermethylation group, the survival rates at 1-, 3-, 5-, and 8-year were 94.16%, 81.71%, 70.78% and 55.93%, respectively. In addition, 3-, 5- and 8-year survival rate in hypermethylation group were significantly higher than that rate in hypomethylation group (Table 2).
Table 2
| Groups | 1-year | 3-year | 5-year | 8-year | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OSR (SE) | P | OSR (SE) | P | OSR (SE) | P | OSR (SE) | P | ||||
| All patients (N=298), n (%) | 0.9292 (0.0149) | 0.7775 (0.0244) | 0.6530 (0.0283) | 0.5121 (0.0381) | |||||||
| Methylation levels, n (%) | 0.274 | 0.026 | 0.007 | 0.013 | |||||||
| Hypomethylation (N=109) | 0.9079 (0.0278) | 0.7080 (0.0442) | 0.5569 (0.0491) | 0.4287 (0.0641) | |||||||
| Hypermethylation (N=189) | 0.9416 (0.0171) | 0.8171 (0.0284) | 0.7078 (0.0339) | 0.5593 (0.0475) | |||||||
OSR, overall survival rate; SE, standard error.
The univariate Cox analysis was performed regarding all variables and MAL methylation (Table 3). Subsequently, the multivariate Cox regression was performed, which included age at diagnosis (P=0.522), gender (P=0.503) and other significant variables revealed in univariate Cox analysis (Table 3). PS was estimated by covariates model including age at diagnosis, gender, preoperative CA19-9, TNM stages, differentiation degree and operation methods. The multivariate Cox regression analysis showed that the preoperative CA19-9, TNM stages, differentiation degree, operation methods and MAL methylation were independent predictors of prognosis for postoperative CRC patients (Table 3, Figure 4). Also, MAL hypermethylation was significantly associated with a favorable clinical outcome with the HR of 0.613 (95% CI: 0.422–0.889, P=0.010) and 0.692 (95% CI: 0.481–0.996, P=0.047) in multivariate Cox regression and PS analysis respectively in all patients (Table 4).
Table 3
| Variables | Number | Univariate Cox | Multivariate Cox | |||||
|---|---|---|---|---|---|---|---|---|
| Patients (N=298) | Deaths (N=123) | Crude HR (95% CI) | P | Adjusted HR (95% CI) | P | |||
| Age at diagnosis | 0.522 | 0.121 | ||||||
| <60 years | 162 | 64 | 1.000 | 1.000 | ||||
| ≥60 years | 136 | 59 | 1.122 (0.788–1.599) | 1.349 (0.924–1.970) | ||||
| Gender | 0.503 | 0.978 | ||||||
| Male | 175 | 70 | 1.000 | 1.000 | ||||
| Female | 123 | 53 | 1.130 (0.791–1.615) | 0.995 (0.673–1.470) | ||||
| CEA | 0.001 | 0.711 | ||||||
| <5 ng/mL | 123 | 36 | 1.000 | 1.000 | ||||
| ≥5 ng/mL | 175 | 87 | 1.935 (1.312–2.855) | 0.915 (0.571–1.466) | ||||
| CA19-9 | 0.000 | 0.000 | ||||||
| <37 U/mL | 217 | 55 | 1.000 | 1.000 | ||||
| ≥37 U/mL | 81 | 68 | 5.628 (3.917–8.084) | 4.756 (3.012–7.511) | ||||
| Tumor location | 0.471 | – | ||||||
| Colon | 110 | 42 | 1.000 | – | ||||
| Rectum | 188 | 81 | 1.147 (0.790–1.665) | – | ||||
| TNM Stages | 0.000 | 0.000 | ||||||
| I–II | 173 | 46 | 1.000 | 1.000 | ||||
| III–IV | 125 | 77 | 3.209 (2.223–4.632) | 2.442 (1.663–3.585) | ||||
| Pathological classification | 0.001 | 0.086 | ||||||
| Prominence | 201 | 69 | 1.000 | 1.000 | ||||
| Others | 97 | 54 | 1.913 (1.312–2.788) | 1.392 (0.954–2.031) | ||||
| Histologic classification | –† | – | ||||||
| Adenocarcinoma | 227 | 95 | 1.000 | – | ||||
| Mucinous adenocarcinoma | 60 | 23 | 0.969 (0.614–1.529) | – | ||||
| Others | 11 | 5 | 1.094 (0.445–2.689) | – | ||||
| Differentiation degree | 0.012 | 0.028 | ||||||
| Poor | 48 | 25 | 1.000 | 1.000 | ||||
| Moderate or well | 250 | 98 | 0.554 (0.350–0.876) | 0.564 (0.338–0.940) | ||||
| Cancer detection methods | 0.281 | – | ||||||
| Symptoms appear | 291 | 119 | 1.000 | – | ||||
| Others | 7 | 4 | 1.731 (0.638–4.694) | – | ||||
| Operation methods | 0.000 | 0.000 | ||||||
| Radical surgery | 285 | 110 | 1.000 | 1.000 | ||||
| Others | 13 | 13 | 17.358 (9.196–32.766) | 6.056 (3.080–11.905) | ||||
| Intestinal stapler | –† | –† | ||||||
| Yes | 223 | 82 | 1.000 | 1.000 | ||||
| No | 63 | 34 | 1.594 (1.062–2.392)* | 1.674 (1.102–2.542)* | ||||
| Unknown | 12 | 7 | 1.575 (0.733–3.384) | 1.179 (0.525–2.649) | ||||
| Intraoperative chemotherapy | –† | – | ||||||
| Yes | 83 | 30 | 1.000 | – | ||||
| No | 200 | 82 | 1.145 (0.751–1.745) | – | ||||
| Unknown | 15 | 11 | 2.220 (1.106–4.458)* | – | ||||
| Postoperative chemotherapy | 0.865 | – | ||||||
| Yes | 135 | 57 | 1.000 | – | ||||
| No | 163 | 66 | 1.031 (0.723–1.471) | – | ||||
| Postoperative radiotherapy | 0.013 | 0.131 | ||||||
| Yes | 14 | 10 | 1.000 | 1.000 | ||||
| No | 284 | 113 | 0.424 (0.215–0.833) | 0.586 (0.293–1.172) | ||||
| MAL methylation | 0.018 | 0.010 | ||||||
| Hypomethylation | 109 | 53 | 1.000 | 1.000 | ||||
| Hypermethylation | 189 | 70 | 0.650 (0.454–0.929) | 0.613 (0.422–0.889) | ||||
†, pooled data was not provided by SPSS 23.0; *, statistically significant association. OS, overall survival; HR, hazard ratio; CI, confidence interval; CEA, carcinoembryonic antigen; TNM, tumor-node-metastasis.
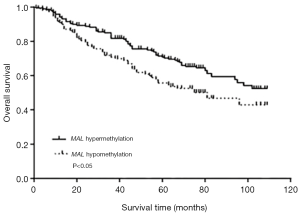
Table 4
| Subgroup | Methylation | Number | Univariate Cox | Multivariate Cox | Propensity score | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | Deaths | Crude HR (95% CI) | P value | Adjusted HR† (95% CI) | P value | Adjusted HR‡ (95% CI) | P value | |||||
| All | 0.018 | 0.010 | 0.047 | |||||||||
| Hypomethylation | 109 | 53 | 1.000 | 1.000 | 1.000 | |||||||
| Hypermethylation | 189 | 70 | 0.650 (0.454–0.929) | 0.613 (0.422–0.889) | 0.692 (0.481–0.996) | |||||||
| Age | ||||||||||||
| <60 years | Hypomethylation | 66 | 33 | 1.000 | 0.006 | 1.000 | 0.001 | 1.000 | 0.013 | |||
| Hypermethylation | 96 | 31 | 0.502 (0.307–0.821) | 0.406 (0.233–0.704) | 0.527 (0.317–0.876) | |||||||
| ≥60 years | Hypomethylation | 43 | 20 | 1.000 | 0.522 | 1.000 | 0.574 | 1.000 | 0.917 | |||
| Hypermethylation | 93 | 39 | 0.838 (0.488–1.439) | 0.851 (0.485–1.493) | 0.968 (0.522–1.796) | |||||||
| Gender | ||||||||||||
| Male | Hypomethylation | 62 | 27 | 1.000 | 0.378 | 1.000 | 0.044 | 1.000 | 0.421 | |||
| Hypermethylation | 113 | 43 | 0.805 (0.497–1.304) | 0.590 (0.353–0.986) | 0.820 (0.506–1.330) | |||||||
| Female | Hypomethylation | 47 | 26 | 1.000 | 0.010 | 1.000 | 0.134 | 1.000 | 0.055 | |||
| Hypermethylation | 76 | 27 | 0.489 (0.285–0.841) | 0.654 (0.375–1.141) | 0.560 (0.309–1.014) | |||||||
| Tumor location | ||||||||||||
| Colon | Hypomethylation | 40 | 20 | 1.000 | 0.031 | 1.000 | 0.008 | 1.000 | 0.024 | |||
| Hypermethylation | 70 | 22 | 0.512 (0.279–0.942) | 0.402 (0.206–0.787) | 0.491 (0.265–0.910) | |||||||
| Rectum | Hypomethylation | 69 | 33 | 1.000 | 0.222 | 1.000 | 0.156 | 1.000 | 0.492 | |||
| Hypermethylation | 119 | 48 | 0.758 (0.487–1.182) | 0.711 (0.443–1.139) | 0.853 (0.541–1.344) | |||||||
| TNM stage | ||||||||||||
| I–II | Hypomethylation | 58 | 20 | 1.000 | 0.083 | 1.000 | 0.409 | 1.000 | 0.086 | |||
| Hypermethylation | 115 | 26 | 0.597 (0.333–1.070) | 0.751 (0.380–1.483) | 0.586 (0.318–1.078) | |||||||
| III–IV | Hypomethylation | 51 | 33 | 1.000 | 0.193 | 1.000 | 0.016 | 1.000 | 0.166 | |||
| Hypermethylation | 74 | 44 | 0.740 (0.470–1.164) | 0.548 (0.336–0.892) | 0.718 (0.449–1.148) | |||||||
†, controlling for the variables which were revealed statistically significant differences in univariate Cox analysis; ‡, covariates model including age at diagnosis, gender, preoperative CA19-9, TNM stages, differentiation degree and operation methods. OS, overall survival; HR, hazard ratio; CI, confidence interval; CEA, carcinoembryonic antigen; TNM, tumor-node-metastasis.
The differences in OS rates stratified by age at diagnosis, gender, tumor location and TNM stages were also analyzed. The CRC MAL hypermethylation patients in subgroup of age at diagnosis <60 and colon cancer had a significantly favorable outcome in the three models (Table 4). In multivariate Cox regression, MAL hypermethylation in male CRC and stages III–IV patients had a favorable outcome significantly with the HR of 0.590 (95% CI: 0.353–0.986, P=0.044) and 0.548 (95% CI: 0.336–0.892, P=0.016), respectively (Table 4).
Discussion
It is well known that biomarkers can be used for disease diagnosis, for predicting disease prognosis and for optimizing treatment strategies. Evidence had shown that methylated DNA signatures can develop into prognostic biomarkers in CRC (5). The MAL gene encodes a non-glycosylated membrane protein that is expressed in a restricted pattern of cell types including T lymphocytes (11), myelin-forming cells (25) and polarized epithelial cells (26). Mal protein is a component of the integral protein machinery for apical transport in polarized epithelial cells (26) and participates in membrane signal transduction (27). Mori et al. reported that MAL promoter hypermethylation was found in primary colon cancer, while the methylation level was significantly lower or absent in normal colonic mucosae (28). However, the implications of MAL methylation on the prognosis of postoperative CRC patients are not clear. To the best our knowledge, this is the first report investigating the association between MAL methylation and prognosis of postoperative CRC patients.
The investigated CpG island of MAL was located in the first exon, which is close to the transcription start position. As a simple, reliable and high sensitive technique, MS-HRM can even assess individual CpG site and detect low-abundance (as low as 0.1–1.0%) methylation (29,30). This technique is more convenient and cost-effective than pyrosequencing, which counts every CpG site (29,31-33). Liu et al. (34), Candiloro et al. (35) and Liu et al. (36) had demonstrated significant consistency of gene methylation between the detection of MS-HRM and pyrosequencing methods.
As a result, we found that the MAL methylation level was significantly higher in tumor tissues than that in adjacent non-tumor tissues. The tumor tissues hypermethylation were found in 76.8% patients, whereas only 4.0% patients in adjacent non-tumor tissues. Our results are in keeping with those from previously published studies (21,37,38). The sensitivity and specificity were 0.768 and 0.960, respectively, with AUC of 0.871 (95% CI: 0.838–0.899) for tumor tissues versus adjacent non-tumor tissues. Lind et al. reported that multi-gene in combination with MAL resulted in a biomarker panel with a sensitivity of 94% and a specificity of 98% (37), which was consistent with the stability and reliability of the results in our study. The high specificity would increase the positive predictive value in judging CRC tumor tissue and adjacent non-tumor tissue.
We collected comprehensive clinicopathologic characteristics data including preoperative CEA, preoperative CA19-9, pathological classification, histologic classification and differentiation degree in our study. The results showed that MAL methylation had no significant correlation with these clinicopathologic characteristics. Lind et al. (37) and Kang et al. (39) also reported that there were no statistically significant differences between MAL methylation and gender, age, tumor location and TNM stages in patients with CRC. In addition, there was also no statistically significant difference between MAL methylation and the clinicopathologic characteristics of patients with gastric cancer (16,20). Unfortunately, the proportion of non-Han population was only 1.0% (3/298), so that the analysis was impossible. Consequently, combined with the above results, MAL methylation might independently affect the survival outcome of CRC patients, rather than affecting the outcome via clinicopathologic characteristics.
Our study is also the first to evaluate the survival rates of CRC patients with different MAL methylation. MAL hypermethylation group had a survival benefit with a longer survival time over 16 months. MAL hypermethylation group had higher OS rates at 3-, 5- and 8-year compared to the hypomethylation group (Table 2) except for 1 year, which indicated that MAL hypermethylation had a significant impact on the long-term survival of postoperative patients with CRC.
Multivariate Cox proportional hazard regression showed that MAL methylation, preoperative CA19-9, TNM stages, differentiation degree and operation methods might be independent prognostic predictors for postoperative patients with CRC. MAL hypermethylated patients had a survival advantage. Usually, as a tumor-suppressor gene, MAL silencing has been associated with promoter hypermethylation, which could re-induce gene expression after demethylation treatment in colon cancer cell lines. In addition, MAL protein was not expressed in the malignant cells (21). Our results were contradicted with the hypothesis that silencing of MAL by hypermethylation is associated with a better prognosis for the CRC patients. Furthermore, other researchers had the same conclusion with ours. For example, Hsi et al. found that Hodgkin lymphoma patients with MAL expression had a poor outcome compared with patients without MAL (40). In addition, Buffart et al. also revealed that the hypermethylation of MAL promoter correlated with a significantly better survival rate of patients with gastric cancer (20). Therefore, we hypothesized that the MAL hypermethylation might lead to additional genetic changes (41) or MAL methylation interact with other gene methylation (42), rather than dependent on MAL methylation alone.
The Cancer Genome Atlas (TCGA) dataset was utilized to validate our results. The CRC datasets of Illumina Human Methylation 450 including colon cancer and rectal cancer were downloaded from UCSC Xena (https://xena.ucsc.edu/) respectively and merged. The dataset included a total of 396 specimens, of which 88 died. The follow-up period ranged from 0.19 to 145.23 months. Only cg22403344 site was located in the same region of our amplification. There was no statistically significant association between methylation of cg22403344 and prognosis of CRC patients, with HR of 1.236 (95% CI: 0.811–1.884, P=0.324). However, with the 20 CG sites in our amplification, there were no similar results as our research.
Our subgroup analysis calculated the differences in OS rates, which was stratified by age at diagnosis, gender, tumor location and TNM stages. First, we found that age at diagnosis <60 years old patients with MAL hypermethylation had a better survival outcome. According to Toyota et al., methylation type A was defined as slight amount methylated in normal mucosa and frequently methylated in tumor tissues, which was associated with aging-specific methylation (41). Based on this, we assumed that MAL methylation might act as type A methylation since they shared same age associated methylation pattern. In addition, we speculated that the association between MAL hypermethylation and a favorable prognosis might be masked in higher age group. Secondly, our result showed that MAL hypermethylation patients had a lower risk of death in colon cancer, whereas difference of OS rates between hypermethylation and hypomethylation was not obvious in rectal cancer group. Thirdly, since patients with early CRC (TNM stages I–II) generally had a better prognosis than those with TNM stages III–IV, we extended the findings that MAL hypermethylation patients with TNM stages III–IV had a higher OS rate. PS has been proved to be a useful, innovative and creative statistical method for evaluating intervention effects in non-experimental or observational studies. The PS analysis included age at diagnosis, gender, preoperative CA19-9, TNM stages, differentiation degree and operation methods into a single covariate, which was used to adjust for baseline differences. The PS analysis confirmed the stability and reliability of the results of multivariate survival analysis.
It is worthy of note that this is a novel study about MAL methylation on the prognosis of postoperative patients with CRC in a patient cohort. We tested tumor tissues and adjacent non-tumor tissues from surgical patients using MS-HRM. Compared with other forms of clinical samples, tissue samples were more stable and reliable. However, there are limitations to be considered. Tumor-specific death was not assessed in our study. The data collected on cancer treatment were limited, and we were restricted from analyzing the association between the MAL methylation and treatment decision, which can be utilized to establish more personalized treatment strategies.
Conclusions
MAL hypermethylation was frequently observed in tumor tissues of patients with CRC and it might be an independent predictor of survival advantage in postoperative patients with CRC. Compared with MAL hypomethylation patients, the hypermethylation patients had a favorable prognosis in younger than 60-year-old patients group and colon cancer patients group.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All patients provided informed consent at the time of sample collection. The study was approved by the Research Ethics Committee of Harbin Medical University.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Arnold M, Sierra MS, Laversanne M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017;66:683-91. [Crossref] [PubMed]
- Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet 2014;383:1490-502. [Crossref] [PubMed]
- DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin 2014;64:252-71. [Crossref] [PubMed]
- Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology 2015;149:1204-25.e12. [Crossref] [PubMed]
- Lopez-Serra L, Esteller M. Proteins that bind methylated DNA and human cancer: reading the wrong words. Br J Cancer 2008;98:1881-5. [Crossref] [PubMed]
- Shima K, Nosho K, Baba Y, et al. Prognostic significance of CDKN2A (p16) promoter methylation and loss of expression in 902 colorectal cancers: Cohort study and literature review. Int J Cancer 2011;128:1080-94. [Crossref] [PubMed]
- Barault L, Charon-Barra C, Jooste V, et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res 2008;68:8541-6. [Crossref] [PubMed]
- Yi JM, Dhir M, Van Neste L, et al. Genomic and epigenomic integration identifies a prognostic signature in colon cancer. Clin Cancer Res 2011;17:1535-45. [Crossref] [PubMed]
- Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet 1999;21:163-7. [Crossref] [PubMed]
- Alonso MA, Weissman SM. cDNA cloning and sequence of MAL, a hydrophobic protein associated with human T-cell differentiation. Proc Natl Acad Sci U S A 1987;84:1997-2001. [Crossref] [PubMed]
- Millán J, Puertollano R, Fan L, et al. The MAL proteolipid is a component of the detergent-insoluble membrane subdomains of human T-lymphocytes. Biochem J 1997;321:247-52. [Crossref] [PubMed]
- Cheong KH, Zacchetti D, Schneeberger EE, et al. VIP17/MAL, a lipid raft-associated protein, is involved in apical transport in MDCK cells. Proc Natl Acad Sci U S A 1999;96:6241-8. [Crossref] [PubMed]
- Puertollano R, Martínez-Menárguez JA, Batista A, et al. An intact dilysine-like motif in the carboxyl terminus of MAL is required for normal apical transport of the influenza virus hemagglutinin cargo protein in epithelial Madin-Darby canine kidney cells. Mol Biol Cell 2001;12:1869-83. [Crossref] [PubMed]
- Martín-Belmonte F, Puertollano R, Millán J, et al. The MAL proteolipid is necessary for the overall apical delivery of membrane proteins in the polarized epithelial Madin-Darby canine kidney and fischer rat thyroid cell lines. Mol Biol Cell 2000;11:2033-45. [Crossref] [PubMed]
- Choi B, Han TS, Min J, et al. MAL and TMEM220 are novel DNA methylation markers in human gastric cancer. Biomarkers 2017;22:35-44. [Crossref] [PubMed]
- Mimori K, Shiraishi T, Mashino K, et al. MAL gene expression in esophageal cancer suppresses motility, invasion and tumorigenicity and enhances apoptosis through the Fas pathway. Oncogene 2003;22:3463-71. [Crossref] [PubMed]
- Overmeer RM, Henken FE, Bierkens M, et al. Repression of MAL tumour suppressor activity by promoter methylation during cervical carcinogenesis. J Pathol 2009;219:327-36. [Crossref] [PubMed]
- Zanotti L, Romani C, Tassone L, et al. MAL gene overexpression as a marker of high-grade serous ovarian carcinoma stem-like cells that predicts chemoresistance and poor prognosis. BMC Cancer 2017;17:366. [Crossref] [PubMed]
- Buffart TE, Overmeer RM, Steenbergen RD, et al. MAL promoter hypermethylation as a novel prognostic marker in gastric cancer. Br J Cancer 2008;99:1802-7. [Crossref] [PubMed]
- Lind GE, Ahlquist T, Kolberg M, et al. Hypermethylated MAL gene—a silent marker of early colon tumorigenesis. J Transl Med 2008;6:13. [Crossref] [PubMed]
- Kalmár A, Péterfia B, Hollósi P, et al. DNA hypermethylation and decreased mRNA expression of MAL, PRIMA1, PTGDR and SFRP1 in colorectal adenoma and cancer. BMC Cancer 2015;15:736. [Crossref] [PubMed]
- Wojdacz TK, Dobrovic A, Hansen LL. Methylation-sensitive high-resolution melting. Nat Protoc 2008;3:1903-8. [Crossref] [PubMed]
- Kalmár A, Péterfia B, Hollósi P, et al. Bisulfite-based DNA methylation analysis from recent and archived formalin-fixed, paraffin embedded colorectal tissue samples. Pathol Oncol Res 2015;21:1149-56. [Crossref] [PubMed]
- Schaeren-Wiemers N, Valenzuela DM, Frank M, et al. Characterization of a rat gene, rMAL, encoding a protein with four hydrophobic domains in central and peripheral myelin. J Neurosci 1995;15:5753-64. [Crossref] [PubMed]
- Puertollano R, Martín-Belmonte F, Millán J, et al. The MAL proteolipid is necessary for normal apical transport and accurate sorting of the influenza virus hemagglutinin in Madin-Darby canine kidney cells. J Cell Biol 1999;145:141-51. [Crossref] [PubMed]
- Marazuela M, Alonso MA. Expression of MAL and MAL2, two elements of the protein machinery for raft-mediated transport, in normal and neoplastic human tissue. Histol Histopathol 2004;19:925-33. [PubMed]
- Mori Y, Cai K, Cheng Y, et al. A genome-wide search identifies epigenetic silencing of somatostatin, tachykinin-1, and 5 other genes in colon cancer. Gastroenterology 2006;131:797-808. [Crossref] [PubMed]
- Wojdacz TK, Dobrovic A. Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res 2007;35:e41. [Crossref] [PubMed]
- Dahl C, Guldberg P. High-resolution melting for accurate assessment of DNA methylation. Clin Chem 2007;53:1877-8. [Crossref] [PubMed]
- White HE, Hall VJ, Cross NC. Methylation-sensitive high-resolution melting-curve analysis of the SNRPN gene as a diagnostic screen for Prader-Willi and Angelman syndromes. Clin Chem 2007;53:1960-2. [Crossref] [PubMed]
- Liu L, Ying C, Zhao Z, et al. Identification of reliable biomarkers of human papillomavirus 16 methylation in cervical lesions based on integration status using high-resolution melting analysis. Clin Epigenetics 2018;10:10. [Crossref] [PubMed]
- Colella S, Shen L, Baggerly KA, et al. Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechniques 2003;35:146-50. [Crossref] [PubMed]
- Liu F, Zhang H, Lu S, et al. Quantitative assessment of gene promoter methylation in non-small cell lung cancer using methylation-sensitive high-resolution melting. Oncol Lett 2018;15:7639-48. [PubMed]
- Candiloro ILM, Mikeska T, Dobrovic A. Assessing alternative base substitutions at primer CpG sites to optimise unbiased PCR amplification of methylated sequences. Clin Epigenetics 2017;9:31. [Crossref] [PubMed]
- Liu Y, Wang Y, Hu F, et al. Multiple gene-specific DNA methylation in blood leukocytes and colorectal cancer risk: a case-control study in China. Oncotarget 2017;8:61239-52. [PubMed]
- Lind GE, Danielsen SA, Ahlquist T, et al. Identification of an epigenetic biomarker panel with high sensitivity and specificity for colorectal cancer and adenomas. Mol Cancer 2011;10:85. [Crossref] [PubMed]
- Sambuudash O, Kim HS, Cho MY. Lack of aberrant methylation in an adjacent area of left-sided colorectal cancer. Yonsei Med J 2017;58:749-55. [Crossref] [PubMed]
- Kang YP, Cao FA, Chang WJ, et al. Gene methylation in stool for the screening of colorectal cancer and pre-malignant lesions. Zhonghua Wei Chang Wai Ke Za Zhi 2011;14:52-6. [PubMed]
- Hsi ED, Sup SJ, Alemany C, et al. MAL is expressed in a subset of Hodgkin lymphoma and identifies a population of patients with poor prognosis. Am J Clin Pathol 2006;125:776-82. [Crossref] [PubMed]
- Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A 1999;96:8681-6. [Crossref] [PubMed]
- Llorente A, de Marco MC, Alonso MA. Caveolin-1 and MAL are located on prostasomes secreted by the prostate cancer PC-3 cell line. J Cell Sci 2004;117:5343-51. [Crossref] [PubMed]


