
Identification of plasma exosomes long non-coding RNA HAGLR and circulating tumor cells as potential prognosis biomarkers in non-small cell lung cancer
Introduction
Non-small cell lung cancer (NSCLC), including squamous-cell carcinoma, adenocarcinoma, and large-cell carcinoma accounts for approx. 80% of all lung cancer cases (1). In spite of broad advances in imaging and therapy, the five-year overall survival for patients with NSCLC remains dismal. Over the recent years, may studies have focused on investigating tumor markers related to NSCLC clinical detection. The improvement of its detection method provides a new direction for early diagnosis, efficacy monitoring and prognosis of tumors.
Circulating tumor cells (CTCs) shed from the primary tumor and enter human peripheral blood. In recent years, many studies have shown that CTCs are an important means of liquid biopsy and play an important role in the diagnosis, treatment evaluation, recurrence and metastasis risk prediction and treatment resistance of many tumor patients, especially in the prognosis of NSCLC (2-6).
Exosome is a microvesicle with the diameter of 30–150 nm that can be secreted by cells (7), which is naturally found in body fluids, including blood, saliva, urine, cerebrospinal fluid and latex. A growing evidence has shown that exosomes are stable and can deliver proteins and nucleic acids to specific cell types, subsequently becoming an important messenger and carrier of tumorigenesis (8,9).
Long non-coding RNA (lncRNAs) are molecules longer than 200 nucleotides in length and with limited or without protein coding function (10,11), which have a key role in the pathogenesis of tumors as well as in other physiological and pathological processes (10). Previous studies have found that lncRNA can be enriched in exosomes (12). Meanwhile, it has been proven that several lncRNAs can directly participate in tumor development and metastasis (13).
lncRNA HAGLR is a 5,090 bp gene located on the antisense strand of a homeobox gene (HOXD) encoding gene on chromosome 2, which belongs to the HOX family. So far, numerous studies have reported that HAGLR is overexpressed in various tumors, such as liver cancer (14), gastric cancer (15) and colorectal cancer (16).
In this study, we examined the expression level of lncRNA HAGLR in plasma exosomes and CTCs of patients and healthy controls by quantitative real-time polymerase chain reaction (qRT-PCR), and initially assessed its possibility as a potential prognosis molecular marker for NSCLC. Meanwhile, the correlations between lncRNA expression level and CTCs and pathological parameters were also analyzed.
Methods
Bioinformatics analysis
GEPIA (http://gepia.cancer-pku.cn/index.html) was used, and The Cancer Genome Atlas (TCGA) was chosen. The tumors were limited to lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). Data screening was performed using the keyword “HAGLR”. The differential expression of HAGLR in LUAD and LUSC was described, respectively. kmplot (http://kmplot.com/analysis/index.php) was used to assess the relationship between the differential expression of HAGLR with clinical prognosis. The correlation between the expression of HAGLR with different stages of NSCLC were elucidated using corresponding statistical methods. All data were calibrated, standardized and converted using log2.
Human sample collection
The 40 patients with NSCLC (case group) who were admitted and treated at The Central Hospital of Enshi Autonomous Prefecture and The Second Xiangya Hospital between August 2017 and December 2018 were included in the study. No patient received adjuvant treatment before radical resection of NSCLC. Lung cell carcinoma was diagnosed according to histopathological diagnosis of the World Health Organization (WHO).
Blood from a total of 8 healthy people who underwent physical examination at the physical examination center in our hospital were collected in the same period. The study protocol was approved by the ethics committee of The Second Xiangya Hospital, Central South University.
Main reagents and instruments
The reagents and instruments included ExoRNeasy Serum/Plasma Maxi Kit (QIAGEN, Germany), chloroform of analytical grade, isopropanol, absolute ethanol (Sinopharm Chemical Reagent Co., Ltd), Primescript TMRTII reverse transcription kit (Takara Biotech, Dalian), PCR primers (Invitrogen), CTC-BIOPSY® abnormal cell separator, optical microscope, ABI 7900 system (Applied Biosystems, Foster City, CA, USA), and ND-1000UV micro-ultraviolet spectrophotometer (Nanodrop, USA).
Plasma separation and CTCs capture
Plasma separation was performed as follows: 4 mL of fasting venous blood from LC patients and healthy subjects was extracted and transferred to EDTA-K2 anticoagulation vacuum tube, which was then centrifuged at 3,000 rpm/min for 15 min at room temperature. The separated plasma was then transferred to a RNase-free EP tube, which was then stored at −80 °C until further use.
CTCs were captured according the following method: 4 mL of fasting venous blood from NSCLC patients and healthy subjects was extracted and transferred to EDTA-K2 anticoagulation vacuum tube, which was rewarmed for 30 min at room temperature. CTCs were enriched and separated using CTC-BIOPSY® abnormal cell separator. Slides were prepared after methylene blue staining for subsequent observation of CTCs detection under a microscope. CTC positive judgment criteria were: (I) karyotype abnormalities, such as nucleus lobulated, mulberry-like, etc.; (II) the nucleoplasm ratio >0.8; (III) cell diameter (long end) >15 µm; (IV) the nucleus was deeply stained and the coloration was uneven, which was mainly because the chromatin of the tumor cells was increased and the particles were coarse; (V) the nuclear membrane was thickened, and there were depressions or wrinkles, making the nuclear membrane irregular; (VI) there were large nucleoli or abnormal nuclear division. CTC was diagnosed if four or more of the above criteria were satisfied.
RNA extraction
Total RNA in plasma exosomes was extracted according to the exoRNeasy Serum/Plasma Maxi Kit, following the manufacturer’s instructions. The extracted total RNA was dissolved in 20 µL of RNase-free water, which was then stored at −80 °C.
qRT-PCR assay lncRNA expression
RNA was reversely transcribed into cDNA according to the Primescript TMRTII kit, following the manufacturer’s instructions. cDNA sample was stored at −80 °C until further use. 18srRNA with stable expression was used as the internal reference, and primers were designed using Primer Premier 5.0 software. The primers were synthesized by Invitrogen after tested by BLAST; the following primer sequences were used: 18srRNA: F 5'-GGACACGGA CAGGATTGACAGA-3', and R 5'-CAGAGTCTCGTTCGTTATCGG A-3'. HAGLR: F: 5'-GGCTCTTCCCTA ATGTGTGG-3', and R 5'-CTCTGGTTGGGTGACTGGTT-3'. The total PCR reaction volume (20 µL) included SYBR Green PCR Master Mix (2×) 10 µL, 1 µL of upstream primers and 1 µL of downstream primers, 2 µL of cDNA template and 6 µL of RNase-Free Water. Parameters in the cycle were: 95 °C for 10 min, 95 °C for 5 s, 58 °C for 25 s and 72 °C for 25 s, a total of 40 cycles; 95 °C for 30 s, 58 °C for 5 s and 95 °C for 30 s. The relative expression of LncRNA was calculated using 2-△△Ct method. △△Ct=△Ct case group (Ct target gene − Ct internal reference gene) − △Ct control group (Ct target gene − Ct internal reference gene).
Statistical analysis
Data analysis was performed using SPSS and Graphpad 6.0 software. The differential expression of lncRNA HAGLR in patients with NSCLC and healthy subjects was analyzed. The correlations of its expression with clinical markers such as gender, age, specific location of tumorigenesis, TNM staging, grading etc., were statistically analyzed.
Results
HAGLR was evidently down-regulated in NSCLC
In this study, using GEPIA platform, LIMMA statistical analysis method was used to examine the expression of HAGLR in NSCLC. Briefly, the cutoff of Log2FC =1 and cutoff of P value = 0.05 were used to match TCGA databases in order to select LUAD and LUSC. This study showed that differentially expressed HAGLR were existed between LUAD or LUSC patients and corresponding adjacent tissues. Briefly, we observed that lower HAGLR expression level in NSCLC patients than adjacent normal tissues (Figure 1, P<0.05).
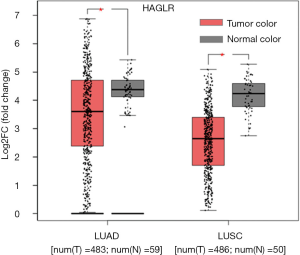
The expression of HAGLR was significant correlation with tumor stage of NSCLC
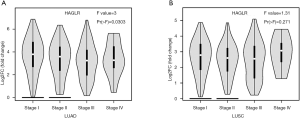
One-way analysis of variance was performed using ANOVA to describe the difference in HAGLR expression at different stages of NSCLC
As shown in (Figure 2), with the staging changes, the expression level of HAGLR is also changing dynamically. Results revealed that the expression of HAGLR was evidently associated with tumor stage of the LUAD (P<0.05), no difference were found for squamous cell carcinoma.
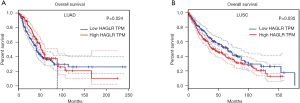
Down-regulation of HAGLR was significant correlation with clinical prognosis in NSCLC
Using GEPIA platform, the correlation between HAGLR expression and the clinical prognosis of NSCLC patients was assessed. Grouping of HAGLR expression was differentiated using median. The survival curve was plotted using KM method, and P value was calculated using Logrank method (Figure 3). the Kaplan-Meier survival plots indicated that the lower expression of HAGLR was associated with poor overall survival in LUAD about 90 months ago, but the trend was reversed after 90 months (Figure 3A, P<0.05). However, down-regulation of HAGLR was significant correlation with good overall survival in LUSC patients (Figure 3B, P<0.05). In summary, the high expression of HAGLR seems to indicate a poor prognosis in patients with NSCLC, especially in patients with squamous cell carcinoma.
The high expression of HAGLR is significantly correlated with the positive rate of CTCs in LUSC
CTCs detection can be used as a predictor of disease recurrence and metastasis in a variety of cancer patients. At present, studies have shown that the more the number of CTCs, the worse the prognosis of patients with NSCLC (6). CTCs were separated and enriched from the peripheral blood of the 40 patients using CTC-BIOPSY® abnormal cell separator. The positive and negative results were determined under the microscope after methylene blue staining (Figure 4). The arrow refers to the CTC after staining one of the patients (Figure 4A), as shown in Figure 4B, other normal cells in the blood. According to the previous survival analysis, High expression HAGLR seems to have a worse survival correlation in squamous cell carcinoma. To confirm the above conclusion again, we used KMplot platform, the Kaplan-Meier survival plots indicated that the higher expression of HAGLR was associated with poor overall survival of LUSC patients (Figure 4C, P<0.05). The same survival trend can be observed in patients with advanced squamous cell carcinoma (Figure 4D). In this study, we further analyzed that the high expression group was associated with a higher positive rate of CTC compared to the HAGLR low expression group of 30 patients with LUSC (Table 1, P<0.05).
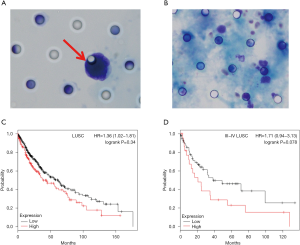
Table 1
| HAGLR expression | Sample size | Tumor stage | CTCs detection rate (%) | χ2 | P value | |||
|---|---|---|---|---|---|---|---|---|
| I–II | III–IV | Negative | Positive | |||||
| Low expression | 13 | 7 | 6 | 6 (46.2) | 7 (53.8) | 4.62 | 0.03 | |
| High expression | 17 | 2 | 15 | 1 (6.00) | 16 (94.0) | |||
CTC, circulating tumor cell; LUSC, lung squamous cell carcinoma.
High expression of HAGLR and high detection rate of CTCs are may be positively correlated with later tumor stage
Pathological data from 40 patients with lung cancer were statistically analyzed. As shown in Table 2, HAGLR high expressions of plasma exosomes were significantly correlated with lymph node metastasis and Tumor stage, but not with gender, age and tumor location.
Table 2
| Clinical indicators | HAGLR expression (%) | χ2 | P value | |
|---|---|---|---|---|
| High expression, N=23 (57.5) | Low expression, N=17 (42.5) | |||
| Gender | 0.051 | 0.822 | ||
| Female | 10 (25.0) | 8 (20.0) | ||
| Male | 13 (32.5) | 9 (22.5) | ||
| Age | 1.52 | 0.218 | ||
| <50 | 9 (22.5) | 10 (25.0) | ||
| ≥50 | 14 (35.0) | 7 (17.5) | ||
| Tumor location | 0.171 | 0.68 | ||
| Left lung | 15 (37.5) | 10 (25.0) | ||
| Right lung | 8 (20.0) | 7 (17.5) | ||
| Lymph node metastasis | 4.181 | 0.041 | ||
| N0 | 5 (12.5) | 9 (22.5) | ||
| N1–3 | 18 (45.0) | 8 (20.0) | ||
| Tumor stage | 5.1 | 0.02 | ||
| Stage I | 2 (5.0) | 4 (10.0) | ||
| Stage II | 4 (10.0) | 6 (15.0) | ||
| Stage III | 8 (20.0) | 5 (12.5) | ||
| Stage IV | 9 (22.5) | 2 (5.0) | ||
Further analysis found that the positive correlation between high CTCs detection rate and the later tumor stage of NSCLC patients was obvious. CTCs were detected in peripheral blood of 28 cases (70.0%) with lung cancer, while no CTCs were detected in healthy subjects. The distribution of detection rate in Tumor stage was: 16.7% (1/6) in stage I, 60.0% (6/10) in stage II, 76.9% (10/13) in stage III and 100% in stage IV (11/11). The obtained results preliminary showed that the detection rate gradually increased (Table 3) and was positively correlated with tumor stage (P<0.05).
Table 3
| Clinical indicators | CTCs detection rate (%) | χ2 | P value | |
|---|---|---|---|---|
| Positive, N=28 (70.0) | Negative, N=12 (30.0) | |||
| Gender | 1.231 | 0.315 | ||
| Female | 11 (27.5) | 7 (17.5) | ||
| Male | 17 (42.5) | 5 (12.5) | ||
| Differentiation | 0.139 | 0.933 | ||
| Low differentiation | 6 (15.0) | 3 (7.5) | ||
| Middle and low differentiation | 11 (27.5) | 5 (12.5) | ||
| Medium differentiation | 11 (27.5) | 4 (10.0) | ||
| Tumor stage | 13.614 | 0.003 | ||
| Stage I | 1 (2.5) | 5 (12.5) | ||
| Stage II | 6 (15.0) | 4 (10.0) | ||
| Stage III | 10 (25.0) | 3 (7.5) | ||
| Stage IV | 11 (27.5) | 0 (0.0) | ||
CTC, circulating tumor cell.
Discussion
In recent years, with the continuous advancement of various detection methods in NSCLC, studies have reported that LncRNAs and CTCs are possible single molecular markers for disease treatment efficacy and prognosis, respectively (17-19). However, the correlation between lncRNA from plasma exosomes and CTCs has less been reported. The combined detection of multiple indicators may be more helpful in the evaluation of curative effect and the monitoring of recurrence and metastasis (20).
lncRNA plays an important role in the development of various tumors. The current published data show that lncRNA is closely related to oncogenes or tumor suppressors and can participate in a variety of cancer processes (such as progression and metastasis) (11). At the same time, the number of CTCs is also related to the tumor stage and prognosis. More CTCs suggest a poor prognosis (21).
In this study, we found that CTCs could be detected in the peripheral blood of NSCLC. In addition, we found that the positive detection rates gradually increased with the tumor stage increase, which was consistent with previous studies (22). Moreover, the correlation analysis results showed that expression of HAGLR in NSCLC plasma exosomes was correlated with lymph node metastasis, Tumor stage, and with the positive detection rate of CTCs, which was consistent with existing reports (23). At the same time, we used bioinformatics analysis and found that high expression of HAGLR in patients with LUSC predicts a worse prognosis. Further analysis of 30 patients with squamous cell carcinoma we collected, the result revealed that patients with high expression of HAGLR had higher detection rates of CTCs, suggesting later tumor stage and worse prognosis. The two conclusions are consistent. However, in patients with adenocarcinoma, the same trend may need to be observed over a longer follow-up period, which may be related to the progression of the patient’s condition in later stages. Further comparison of the survival curves reveals that the horizontal axis shows that the overall survival of patients with squamous cell carcinoma is shorter than that of adenocarcinoma. This may suggest that patients with advanced squamous cell carcinoma are predominant, leading to differences in survival in different histopathological types. With the extension of follow-up time, with the 90-month node, it is possible that adenocarcinoma patients with some disease progression can see a trend consistent with squamous cell carcinoma. That is, the low expression of HAGLR has a better prognosis. This does require an expansion of the sample size in subsequent studies and validation based on different stages and stratified analysis of different pathological types. In this study, there is a different concept of grouping that needs to be clarified. We used a public database to analyze the expression of HAGLR in LUAD or LUSC. The control group was normal tissue. The analysis of the expression of HAGLR in our collected clinical samples was related to the clinicopathological features, all of which were the expression of tumor samples, and the median was used to distinguish between high and low expression.
HOXD cluster antisense RNA1 (HAGLR) is located between the HOXD1 gene and the HOXD3 gene. It has eight exons, and the opposite transcription direction (24). The biological functions and mechanisms of lncRNA HAGLR in different human tumors have been reported, including bladder cancer, cervical cancer, gastric cancer, ovarian cancer, glioma, hepatocellular carcinoma and NSCLC. Lu et al. (25) have confirmed that the expression level of HAGLR significantly increases in lung cancer tissues and lung cancer cell lines. In addition, the correlation analysis has shown that the expression of HAGLR is closely related to tumor stage, lymph node metastasis and survival status. In this study, plasma exosomes lncRNA HAGLR was extracted. Our data suggested that the correlation between clinical stages and lymph node metastasis was consistent with tissue level. They also showed a good clinical prospect in liquid biopsy. HAGLR expression could be detected in peripheral blood while detecting CTC and the detection rate significantly increased with the increase in tumor stages.
In conclusion, plasma exosomal lncRNA detection and CTCs detection are a non-invasive, rapid and real-time method. The correlation between the abnormally expressed HAGLR and CTCs detection rate may be an important molecular marker for the disease progression and possible metastatic risk prediction of NSCLC, and may have more effective predictive value than traditional single molecular markers.
However, in subsequent studies, the standardization of exosome extraction methods and the effective isolation of exosomes are areas for improvement, and the need to expand the sample size for further stratification analysis in LUAD and LUSC, Clinical data related to more clinical pathological features and survival prognosis were obtained to verify the clinical value of the study.
Conclusions
In summary, we revealed that high expression of plasma exosome HAGLR is associated with a high detection rate of CTCs in NSCLC patients. Moreover, the high expression of HAGLR shows a poor prognosis in LUSC and adenocarcinoma patients whose condition may progress. A larger number of CTCs suggest that the disease may progress in line with the previous conclusions. The study provides a new idea that plasma exosomes HAGLR and CTCs may be predictors of progression of NSCLC.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.43). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the ethics committee of The Second Xiangya Hospital, Central South University. Individual informed consent was waived due to the nature of the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Traynor AM, Schiller JH. Systemic treatment of advanced non-small cell lung cancer. Drugs Today (Barc) 2004;40:697-710. [Crossref] [PubMed]
- Bahnassy AA, Salem S E, Mohanad M, et al. Prognostic significance of circulating tumor cells (CTCs) in Egyptian non-metastatic colorectal cancer patients: A comparative study for four different techniques of detection (Flowcytometry, Cell Search, Quantitative Real-time PCR and Cytomorphology). Exp Mol Pathol 2019;106:90-101. [Crossref] [PubMed]
- Pawlikowska P, Faugeroux V, Oulhen M, et al. Circulating tumor cells (CTCs) for the noninvasive monitoring and personalization of non-small cell lung cancer (NSCLC) therapies. J Thorac Dis 2019;11:S45. [Crossref] [PubMed]
- Cristofanilli M, Pierga J Y, Reuben J, et al. The clinical use of circulating tumor cells (CTCs) enumeration for staging of metastatic breast cancer (MBC): international expert consensus paper. Crit Rev Oncol Hematol 2019;134:39-45. [Crossref] [PubMed]
- Mamdouhi T, Twomey JD, McSweeney KM, et al. Fugitives on the run: circulating tumor cells (CTCs) in metastatic diseases. Cancer Metastasis Rev 2019;38:297-305. [Crossref] [PubMed]
- Normanno N, De Luca A, Gallo M, et al. The prognostic role of circulating tumor cells in lung cancer. Expert Rev Anticancer Ther 2016;16:859-67. [Crossref] [PubMed]
- Whiteside TL. The emerging role of plasma exosomes in diagnosis, prognosis and therapies of patients with cancer. Contemp Oncol (Pozn) 2018;22:38-40. [Crossref] [PubMed]
- Regev-Rudzki N, Wilson DW, Carvalho TG, et al. Cell-cell communication between malaria-infected red blood cells via exosome-like vesicles. Cell 2013;153:1120-33. [Crossref] [PubMed]
- Sun N, Sun SG, Lu Z L, et al. Diagnostic value of protein markers in plasma exosomes of lung squamous cell carcinoma. Zhonghua Zhong Liu Za Zhi 2018;40:418-21. [PubMed]
- Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci 2016;73:2491-509. [Crossref] [PubMed]
- Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018;172:393-407. [Crossref] [PubMed]
- Conigliaro A, Costa V, Dico AL, et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer 2015;14:155. [Crossref] [PubMed]
- Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol 2011;21:354-61. [Crossref] [PubMed]
- Wang H, Huo X, Yang XR, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer 2017;16:136. [Crossref] [PubMed]
- Zheng L, Chen J, Zhou Z, et al. Knockdown of long non-coding RNA HOXD-AS1 inhibits gastric cancer cell growth via inactivating the JAK2/STAT3 pathway. Tumour Biol 2017;39:1010428317705335. [Crossref] [PubMed]
- Li X, Zhao X, Yang B, et al. Long non-coding RNA HOXD-AS1 promotes tumor progression and predicts poor prognosis in colorectal cancer. Int J Oncol 2018;53:21-32. [PubMed]
- Tian X, Xu G . Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: systematic review and meta-analysis. BMJ Open 2015;5:e008653. [Crossref] [PubMed]
- Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. Int J Cancer 2017;140:1955-67. [Crossref] [PubMed]
- Wei T, Zhu D, Yang Y, et al. The application of nano-enrichment in CTC detection and the clinical significance of CTCs in non-small cell lung cancer (NSCLC) treatment. PLoS One 2019;14:e0219129. [Crossref] [PubMed]
- Alama A, Coco S, Genova C, et al. Prognostic Relevance of Circulating Tumor Cells and Circulating Cell-Free DNA Association in Metastatic Non-Small Cell Lung Cancer Treated with Nivolumab. J Clin Med 2019;8:1011. [Crossref] [PubMed]
- Dong J, Zhu D, Tang X, et al. Circulating tumor cells in pulmonary vein and peripheral arterial provide a metric for PDL1 diagnosis and prognosis of patients with non small cell lung cancer. PLoS One 2019;14:e0220306. [Crossref] [PubMed]
- Zhang Y, Zheng H, Zhan Y, et al. Detection and application of circulating tumor cell and circulating tumor DNA in the non-small cell lung cancer. Am J Cancer Res 2018;8:2377-86. [PubMed]
- Xia H, Jing H, Li Y, et al. Long noncoding RNA HOXD-AS1 promotes non-small cell lung cancer migration and invasion through regulating miR-133b/MMP9 axis. Biomed Pharmacother 2018;106:156-62. [Crossref] [PubMed]
- Yarmishyn AA, Batagov AO, Tan JZ, et al. HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome. BMC Genomics 2014;15:S7. [Crossref] [PubMed]
- Lu C, Ma J, Cai D. Increased HAGLR expression promotes non-small cell lung cancer proliferation and invasion via enhanced de novo lipogenesis. Tumour Biol 2017;39:1010428317697574. [Crossref] [PubMed]


