
Effect of NOP2 knockdown on colon cancer cell proliferation, migration, and invasion
Introduction
As the third most prevalent cancer type, colon cancer is also the fourth leading cause of cancer-related deaths worldwide. More than 1.2 million people are diagnosed with colon cancer, and >600,000 die from the disease every year (1-3). Additionally, colon cancer is the fifth leading cause of death in China, constituting a major health problem (4). Advanced age, alcohol consumption, obesity, smoking, lack of physical activity, high consumption of red or processed meat, low calcium intake, and very low consumption of fruits and vegetables may contribute to colon cancer (5). In terms of genes, activation of proto-oncogenes and inactivation of tumor suppressor genes may lead to colon cancer (6). Several oncogenes and tumor suppressor genes, such as Adenomatous polyposis coli, Tumor protein 53, and KRAS proto-oncogene GTPase have been identified as the key factors in tumor formation years ago (7-10). However, although we have an improved understanding of the development and progression of colon cancer, a cure is yet to be found.
Nucleolar protein p120 (NOP2) expression has been shown to be significantly upregulated in most types of human malignant tumor cells, such as breast cancer cells, but not in healthy, resting cells (11-13). NOP2 has been identified as a “telomerase RNA component” (TERC)-binding protein, and it can also bind to the “T-cell factor” (TCF)-binding element of the Cyclin D1 promoter, activating the transcription of Cyclin D1 (14). Although NOP2 could be implicated as a tumor cell maker, it has not been specifically studied in colon cancer.
In the study presented herein, we knocked down NOP2 in the human colon cancer cell lines, LOVO and HCT116, via siRNA transfection to determine the role of NOP2 in colon cancer. The effects of NOP2 silencing on the colon cancer cells were evaluated by CCK-8 and transwell assays as well as transcriptome sequencing.
Methods
Cell culture
Colon cancer cells (HCT116 and LOVO) were obtained from Sangon Biotech (Shanghai, China). HCT116 cell lines were cultured in F12K medium (GIBCO, CA, USA) supplemented with 10% FBS and 1% Penicillin/Streptomycin (Sangon Biotech, Shanghai, China). LOVO cell lines were cultured in RPMI-1640 medium (CORNING, NY, USA) supplemented with 10% FBS and 1% Penicillin/Streptomycin (Sangon Biotech, Shanghai, China). Cells were grown at 37 °C in a humidified atmosphere with 5% CO2.
Fluorescence quantitative PCR
Total RNA was extracted from cultured cells using the TRIZOL reagent (Invitrogen, CA, USA) according to the manufacturer’s protocol. The RNA concentration and quality were determined by OD260/OD280 and agarose gel electrophoresis. Total RNA (1 µg) was then reverse-transcribed with a commercial reverse transcription kit (Thermo Bio.), and then PCR was performed with the 2× Master Mix kit (Roche) according to the manufacturer’s instructions. FastStart Universal SYBR Green Master mix was used to amplify cDNA on a QuantStudio 6 Flex Real-Time PCR System (ThermoFisher, Shanghai, China). The primers used for RT-PCR were as follows:
NOP2-1 forward, 5'-TGTCTGAGCTGGTGGAGTTCTTAG-3′ and reverse, 5'-ACCCCACGATTGATTAGAGCC-3'; GAPDH forward, 5'-AGAAGGCTGGGGCTCATT-3' and reverse, 5'- TGCTAAGCAGTTGGTGGTG-3'.
The relative expression levels were evaluated by the 2−ΔΔct method using GAPDH as the internal control for normalization of the values.
siRNA transfection
LOVO and HCT116 cells were seeded at a density of 3×105 per well on 6-well plates and grown for 24 h in the complete medium until 80–90% of confluency. Then, the cells were rinsed with fresh medium, incubated with siRNA-NOP2 (GenePharma, Shanghai, China) and LipofectamineTM2000 (Invitrogen, CA, USA), which had been diluted beforehand by OPTI-MEM (CORNING, NY, USA). The sequences of the siRNAs are listed in Table 1.
Table 1
| RNA oligo | Sequences (5' to 3') |
|---|---|
| NOP2-homo-2215 | GCAACGAUCACCUAAAUUATT |
| UAAUUUAGGUGAUCGUUGCTT | |
| NOP2-homo-1836 | CCCACAAUAUGGAUGGGUUTT |
| AACCCAUCCAUAUUGUGGGTT | |
| NOP2-homo-338 | GCCAAACCAUUGCCUGGAATT |
| UUCCAGGCAAUGGUUUGGCTT | |
| NC | UUCUCCGAACGUGUCACGUTT |
| ACGUGACACGUUCGGAGAATT |
NOP2, nucleolar protein p120; NC, negative control.
Western blot analysis
After analyzing the cells in RIPA buffer (Thermo Fisher Scientific, Waltham, MA), total protein concentration was measured using the BCA assay kit (Pierce Biotechnology, Inc., Rockford, IL, USA). The same quantity of total proteins from each sample was resolved by 10% SDS-PAGE and then electrophoretically transferred onto polyvinylidene fluoride (Sangon Biotech, Shanghai, China) membranes at 80 V for 2 h. The membranes were blocked with TBST solution (0.1% Tween20) containing 5% skim milk and incubated with the primary antibodies anti-NOP2 (1:500; 10448-1-AP, Proteintech, Wuhan, China) and anti-GAPDH (1:1,000; 60004-1-Lg, Proteintech, Wuhan, China) at 4 °C overnight. The membranes were washed with TBST and incubated with the HRP-conjugated goat anti-mouse IgG (1:1,000, A0216, Beyotime, Shanghai, China) or goat anti-rabbit IgG (1:5,000, Ab721, Abcam) for 2 h. The membranes were then washed with 1% TBST for 3 times, and the protein bands were visualized using ECL (Thermo Fisher Scientific, Waltham, MA, USA) and a ChemiDoc MP system.
CCK-8 assay
Cells were seeded in a 96-well plate (3599, CORNING, NY, USA) at a density of 2×103 per well. Each plate contained 6 replicates, and the marginal wells were added sterile water for the contrast. After culturing for 0, 24, 48, 72, or 96 h, CCK-8 (Beyotime, Shanghai, China) was added and OD450 of the culture was measured.
Transwell assay
Cells were washed with an appropriate amount of 1× PBS, and then an appropriate amount of 0.25% Trypsin-EDTA (25200072, GIBCO, CA, USA) was added. The culture vessel was gently shaken to submerge all the cells and returned to the incubator for 3 min. After the cells retracted and rounded up, complete medium at the same amount used for the culture was added to terminate the tryptic digestion. The plates were then centrifuged and the supernatant was removed. The cells were resuspended in serum-free RPMI-1640 medium (CORNING, NY, USA) and counted. Their concentration was adjusted to 2×105/mL. Then, 500 µL of the cell suspension was loaded into the upper chambers (CORNING, NY, USA) of the transwell device, and 700 µL complete medium with 10% FBS was added into the lower chamber . After 24 h of culture, the cells underneath the transwell membrane were stained and counted under a microscope by evaluating three random high-power fields. For the transwell and invasion experiments, 24-well plates with 8.0-µm transparent PET membrane (353097, BD Falcon, NY, USA), and Corning BioCoat™ Matrigel® invasion chambers with 8.0-µm PET membrane in 24-well plates were used, respectively.
Transcriptome sequencing
Total RNA was extracted from cultured cells using the TRIZOL reagent (Invitrogen, CA, USA) according to the manufacturer's protocol. Oligo-dT magnetic beads were used for mRNA isolation. The isolated mRNA was subjected to mRNA fragmentation in a fragmentation buffer by divalent cations and under high-temperature conditions. Single-strand cDNAs from the fragmented mRNAs were synthesized using reverse transcriptase and random hexamer primers. The mRNA strands of the heteroduplexes were digested by RNase H to synthesize the complementary strands of the cDNAs, thereby realizing cDNA second strand synthesis. The sticky ends of the double-stranded cDNAs are blunted by the addition of End Repair Mix. An Adenosine monophosphate was added to the 3' ends of the double-stranded cDNAs for ligation of the cDNAs to the Y-shaped linker. The adaptor-ligated library was size-selected using Ampure Beads (Beckman, Brea, California, USA). The library was PCR-amplified for 15 cycles, and then purified using Ampure Beads (Beckman, Brea, CA, USA) and quantified with the Quant-iT™ PicoGreen® dsDNA Assay Kit (Life, Carlsbad, CA, USA) following the manufacturer's protocol. The sizes of the DNA fragments were determined using Agilent 2200 (Agilent, Santa Rosa, CA, USA). Sequencing was performed on the Illumina sequencing platform (Illumina, San Diego, CA, USA). All the raw data was converted to the Fastq format. The differentially expressed genes were analyzed by DEGSeq algorithm, and Gene Ontology (GO) and pathway analysis of significant differential genes were performed by Fisher.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 6. Significance of differences between two groups was determined using the Student’s t-test. Correlation between the NOP2 expression and characteristics of the colon cancer patients was evaluated by using the Chi-square test. P<0.05 was defined as the significance level.
Results
NOP2 expression was significantly upregulated in colon cancer cells
Fluorescence quantitative PCR was used to investigate the expression of NOP2 in colon cancer tissues and cell lines. As shown in Figure 1A,B, mRNA and protein levels of NOP2 were significantly upregulated in colon tumor samples compared with those in the healthy controls. Similarly, NOP2 was upregulated in the colon cancer cell lines examined relative to the non-transformed cells (Figure 1C,D). Moreover, expression of NOP2 significantly showed a positive correlation with lymphatic metastasis and high TNM stage (Table 2).
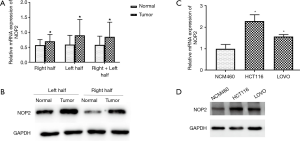
Table 2
| Features | Number | NOP2 expression | P value | |
|---|---|---|---|---|
| High | Low | |||
| Gender | 0.691 | |||
| Male | 13 | 8 | 5 | |
| Female | 13 | 7 | 6 | |
| Age | 0.778 | |||
| <60 | 5 | 3 | 2 | |
| ≥60 | 21 | 14 | 7 | |
| Tumor size | 0.169 | |||
| <5 | 20 | 13 | 7 | |
| ≥5 | 6 | 2 | 4 | |
| Differentiation | 0.42 | |||
| Moderate | 8 | 4 | 4 | |
| Poor | 18 | 12 | 6 | |
| Lymphatic metastasis | 0.006 | |||
| Y | 16 | 12 | 4 | |
| N | 10 | 2 | 8 | |
| TNM stage | 0.012 | |||
| I and II | 11 | 3 | 8 | |
| III and IV | 15 | 11 | 4 | |
NOP2, nucleolar protein p120.
NOP2 was efficiently silenced in colon cancer cell lines
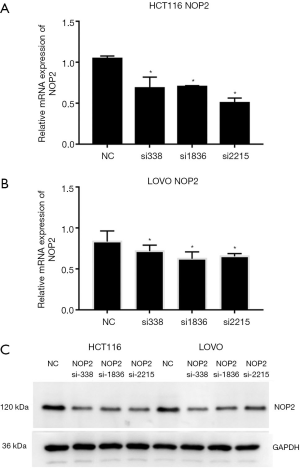
After siRNA infection, we used fluorescence quantitative PCR and western blot analysis to examine the expression of NOP2 in both HCT116 and LOVO cell lines. Fluorescence quantitative PCR results in Figure 2A,B showed that all the three siRNAs efficiently knocked down NOP2 in both HCT116 and LOVO cell lines. Western blot analysis demonstrated that NOP2 protein levels were also reduced (Figure 2C). The results indicated that the most effective siRNA among the three tested siRNAs was si2215, and thus the subsequent experiments were performed with this siRNA.
Silencing NOP2 inhibited colon cancer cell proliferation, invasion, and migration
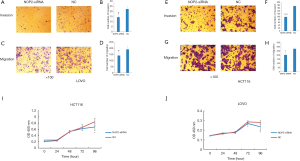
The effect of NOP2 silencing on colon cancer cells was explored by CCK-8 and Transwell assays. As indicated in Figure 3A,B,C,D,E,F,G,H,I,J, the proliferation (P<0.05, P<0.05), invasion (P<0.0005, P<0.05), and migration (P<0.005, P<0.05) of the colon cancer cells (HCT116 and LOVO, respectively) were significantly suppressed in the NOP2 knockdown groups compared with those in the negative control (NC) groups.
Differentially expressed genes between the NOP2 knockdown and NC groups
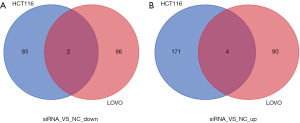
As shown in Figure 4A,B, 88 and 94 genes were significantly down- and upregulated, respectively, in LOVO cells upon NOP2 knockdown, relative to those in the control group (|log2 Fold Change| >1, FDR <0.05). In the NOP2-knockdown HCT116 cells, 95 and 175 genes were down- and upregulated, respectively, relative to those in the control group (|log2 Fold Change| >1, FDR <0.05). Among all these genes, ASMTL and C6orf52 were significantly downregulated in both the cell lines with NOP2 knockdown, while MUC19, TXK, APOBEC2, and RBM44 were significantly upregulated.
GO and pathway analysis of the NOP2-knockdown colon cancer cell lines
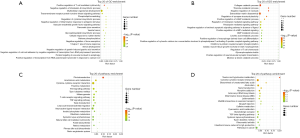
To study the function of NOP2 in colon cancer and the molecular mechanisms involved, GO and pathway analysis were performed for differentially expressed genes induced by NOP2 knockdown. The roles of NOP1 were investigated by GO analysis within three different domains (BP, CC, and MF). All the terms that showed significant differences, ranked by −log10 (P value), are displayed in Figure 5A (HCT116) and 5B (LOVO). Top terms for HCT116 cell line included positive regulation of T cell-mediated cytotoxicity, negative regulation of interleukin-6 biosynthetic process, and multicellular organismal development. For the LOVO cell line, top terms included collagen catabolic process, thiamine metabolism, and negative regulation of cytolysis. The top 20 significantly enriched KEGG pathways, ranked by −log10 (P value), are displayed in Figure 5C (LOVO) and 5D (HCT116). The identified pathways in both the cell lines were Cytokine-cytokine receptor interaction, type I diabetes mellitus, taste transduction, and systemic lupus erythematosus.
Discussion
Colon cancer is one of the leading causes of cancer-related death, and the incidence of colon cancer has recently been increasing (15,16). There are more than one million new colon cancer cases and approximately 700,000 people die from colon cancer each year (17). The etiology of colon cancer is multifactorial and has remained poorly defined (18).
NOP2, as an important regulator of the cell cycle, has been shown to be associated with a non-canonical mechanism by which telomerase promotes cell proliferation in breast cancer cells (14). However, the role of NOP2 in colon cancer remains unclear. We were stimulated to investigate this role by the current lack of clarity. Our results demonstrated that silencing NOP2 suppressed colon cancer cell proliferation, invasion, and migration. Furthermore, we also found that the levels of several genes were significantly different between the experimental and control groups.
NOP2 is a nucleolar antigen expressed in proliferating cells and many human tumors (19). Translational repression of NOP2 has been shown to suppress breast cancer proliferation (20). NOP2 also plays an important role in the proliferation of gliomas, and the ribozyme against NOP2 mRNA can suppress glioma cell growth (21). Similar conclusions have also been drawn in the prostate (22) and lung cancer (23). Our results here showed that NOP2 was significantly overexpressed in neoplastic cells, in line with the results of a previous study (24). To further understand the role of NOP2 in colon cancer, we transfected colon cancer cells with NOP2-siRNAs and found that, in addition to cell proliferation, NOP2 also had a role in cell migration and invasion. We demonstrated that silencing NOP2 significantly suppressed colon cancer cell proliferation, migration, and invasion.
Furthermore, we performed transcriptome sequencing to explore the action mechanism of NOP2. We thereby found that TXK tyrosine kinase was significantly upregulated in the NOP2 knockdown groups. As a member of the Tec family of tyrosine kinases, TXK is the most commonly found growth factor receptor in many tumors and has a key signal-transducing role (25,26). It has been shown to have anti-cancer/immunomodulatory effects (27). Accordingly, our results show a plausible action mechanism of NOP2 in colon cancer, requiring further research.
Conclusions
In conclusion, our study demonstrates that lack of NOP2 significantly downregulates ASMTL and C6orf52, while upregulating MUC19, TXK, APOBEC2, and RBM44. Reducing NOP2 levels significantly suppresses colon cancer cell proliferation, migration, and invasion. This effect may be related to differential expression of other genes, such as TXK, and this correlation needs to be elaborated in future research.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.46). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional ethical approval and individual informed consent were waived due to the nature of the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Song L, Fan Z, Jun N, et al. Tumor specific delivery and therapy mediate by integrin beta6-target immunoliposomes for beta6-siRNA in colon carcinoma. Oncotarget 2016;7:85163-75. [Crossref] [PubMed]
- Loganathan L, Muthusamy K. Current Scenario in Structure and Ligand-Based Drug Design on Anti-colon Cancer Drugs. Curr Pharm Des 2018;24:3829-41. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- American Cancer Society. Cancer Facts & Figures. American Cancer Society: Atlanta, GA, USA, 2016.
- Zhou D, Yang L, Zheng L, et al. Exome capture sequencing of adenoma reveals Genetic alterations in multiple cellular pathways at the early stage of colorectal tumorigenesis. PLoS One 2013;8:e53310. [Crossref] [PubMed]
- Korinek V, Barker N, Morin PJ, et al. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC-/- colon carcinoma. Science 1997;275:1784-7. [Crossref] [PubMed]
- Morin PJ, Sparks AB, Korinek V, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 1997;275:1787-90. [Crossref] [PubMed]
- Baker SJ, Markowitz S, Fearon ER, et al. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 1990;249:912-5. [Crossref] [PubMed]
- Aoki Y, Niihori T, Narumi Y, et al. The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum Mutat 2008;29:992-1006. [Crossref] [PubMed]
- Ochs RL, Reilly MT, Freeman JW, et al. Intranucleolar localization of human proliferating cell nucleolar antigen p120. Cancer Res 1988;48:6523-9. [PubMed]
- Jhiang SM, Yaneva M, Busch H. Expression of human proliferation-associated nucleolar antigen p120. Cell Growth Differ 1990;1:319-24. [PubMed]
- Fonagy A, Swiderski C, Dunn M, et al. Antisense-mediated specific inhibition of P120 protein expression prevents G1- to S-phase transition. Cancer Res 1992;52:5250-6. [PubMed]
- Hong J, Lee JH, Chung IK. Telomerase activates transcription of cyclin D1 gene through an interaction with NOL1. J Cell Sci 2016;129:1566-79. [Crossref] [PubMed]
- Birt DF, Phillips GJ. Diet, genes, and microbes: complexities of colon cancer prevention. Toxicol Pathol 2014;42:182-8. [Crossref] [PubMed]
- Calvert PM, Frucht H. The genetics of colorectal cancer. Ann Intern Med 2002;137:603-12. [Crossref] [PubMed]
- Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg 2009;22:191-7. [Crossref] [PubMed]
- Zhai H, Shi Y, Chen X, et al. CacyBP/SIP promotes the proliferation of colon cancer cells. PLoS One 2017;12:e0169959. [Crossref] [PubMed]
- Freeman JW, Busch RK, Gyorkey F, et al. Identification and characterization of a human proliferation-associated nucleolar antigen with a molecular weight of 120,000 expressed in early G1 phase. Cancer Res 1988;48:1244-51. [PubMed]
- Lewinska A, Bednarz D, Adamczyk-Grochala J, et al. Phytochemical-induced nucleolar stress results in the inhibition of breast cancer cell proliferation. Redox Biol 2017;12:469-82. [Crossref] [PubMed]
- Sato K, Nishi T, Takeshima H, et al. Expression of p120 nucleolar proliferating antigen in human gliomas and growth suppression of glioma cells by p120 ribozyme vector. Int J Oncol 1999;14:417-24. [PubMed]
- Saijo Y, Sato G, Usui K, et al. Expression of nucleolar protein p120 predicts poor prognosis in patients with stage I lung adenocarcinoma. Ann Oncol 2001;12:1121-5. [Crossref] [PubMed]
- Bantis A, Giannopoulos A, Gonidi M, et al. Expression of p120, Ki-67 and PCNA as proliferation biomarkers in imprint smears of prostate carcinoma and their prognostic value. Cytopathology 2004;15:25-31. [Crossref] [PubMed]
- Ueki T, Nakayama Y, Sugao Y, et al. Significance of the expression of proliferation-associated nucleolar antigen p120 in human colorectal tumors. Hum Pathol 1997;28:74-9. [Crossref] [PubMed]
- Mihara S, Suzuki N. Role of Txk, a member of the Tec family of tyrosine kinases, in immune-inflammatory diseases. Int Rev Immunol 2007;26:333-48. [Crossref] [PubMed]
- Zhang Y, Wester L, He J, et al. IGF1R signaling drives antiestrogen resistance through PAK2/PIX activation in luminal breast cancer. Oncogene 2018;37:1869-84. [Crossref] [PubMed]
- Tavares TS, Nanus D, Yang XJ, et al. Gene microarray analysis of human renal cell carcinoma: the effects of HDAC inhibition and retinoid treatment. Cancer Biol Ther 2008;7:1607-18. [Crossref] [PubMed]


