
Withaferin A suppresses skin tumor promotion by inhibiting proteasome-dependent isocitrate dehydrogenase 1 degradation
Introduction
A metabolic shift often accompanies carcinogenesis. Cancer cells lean to utilizing a high rate of glycolysis to generate energy instead of oxidative phosphorylation (OXPHOS) even in the presence of oxygen, known as the “Warburg Effect” (1) or aerobic glycolysis. An important benefit of enhanced aerobic glycolysis is that the largely produced metabolic intermediates serve as the building blocks for biosynthesis (2). Associated with the metabolic switch is the dysregulation of crucial metabolic enzymes. Isocitrate dehydrogenases (IDHs) as the most pivotal metabolic enzyme catalyze the nicotinamide adenine dinucleotide (NAD+)-dependent or nicotinamide adenine dinucleotide phosphate (NADP+)-dependent transformation of isocitrate to α-ketoglutarate (3). IDHs possess three isoforms: IDH3 is the main form of IDH involved in the TCA cycle, and no mutation in any of the IDH3 subunits is reported in cancer cells at present. IDH1 and IDH2 mutations have been reported in gliomas and acute myelogenous leukemia (4). Mutant IDHs produce 2-hydroxyglutarate. It is possible to regulate the hypermethylation of RIP3 promotor mediated by DNA methyltransferase (DNMT1), impairing necroptosis to contribute to tumorigenesis (5); therefore, 2-hydroxyglutarate has been recognized as an onco-metabolite (6). Unlike IDH2/3, IDH1 located in cytosol, whose substrate and product could shuttle between mitochondria and cytosol, has been determined as a tumor suppressor since its stability plays an essential role in carcinogenesis (7). Our previous study demonstrated that the protein and activity levels of IDH1 were inhibited by tumor promoter treatment, not IDH2. Therefore, maintaining the expression and activity levels of IDH1 would be of importance to carcinogenesis inhibiting.
As one of the major bioactive compounds derived from Withania somnifera, Withaferin A (WA) has been used for the treatment of nervous and sexual disorders (8). Recent studies have demonstrated the anti-tumor potential of WA, which shows that WA exerts the anti-proliferation activity in non-small cell lung cancers (9), osteosarcoma (10), breast cancer (11), prostate cancer (12), colon cancer (13), and pancreatic cancer (14), either in vitro or in vivo. Meanwhile, a clinical trial of WA has been launched for treatment of metastatic melanoma (15). Our early studies have shown that WA inhibits skin cell transformation and blocks tumor promoter-induced metabolic alterations (16), which make WA a potential chemopreventive agent. However, the molecular mechanisms of chemoprevention could be different from its anti-tumor activities.
Increasing evidences show that ubiquitin-proteasome pathway (UPP) inducing degradation of tumor suppressors plays a contributing role during carcinogenesis (17,18). In detail, to cooperate with ubiquitin-activating enzymes E1 and ubiquitin-conjugating enzymes E2, ubiquitin E3 ligases combine and modify specific substrates leading to their degradation by 26S proteasome (19). This ubiquitin E3 ligase pathway is crucial during tumorigenesis (20,21), which may also serve as the target for chemoprevention. Based on computer simulation (22) and experimental data (17,23), it has been proved that WA could bind to the 20S, 26S proteasome core complex, to inhibit the UPP. In present study, we will examine whether WA may target UPP, thereby stabilize IDH1 as an important mechanism of chemoprevention.
Methods
Cell lines, reagents, and treatment
Murine skin epidermal JB6 Cl-41 P+ cells (purchased from American Type Culture Collection) were grown in EMEM (Gibco) medium containing 5% fetal bovine serum (FBS), penicillin and streptomycin in a 37 °C incubator under 5% CO2. WA, dissolved in dimethyl sulfoxide (DMSO; Sigma, St Louis, MO, USA), was purchased from ChromaDex (ASB-00023250; Irvine, CA, USA). 12-O-tetradecanoylphorbol-13-acetate (TPA; P1585, Sigma), MG-132 (HY-13259; MCE, Monmouth Junction, NJ, USA), and cycloheximide (CHX; C7698, Sigma) were dissolved in DMSO.
Isolation of total RNA and real-time PCR
Total RNA was isolated from cells using E.Z.N.A total RNA kit II (OMEGA, Norcross, GA, USA) following the manufacturer’s instruction. Two µg of the purified RNA was reverse transcribed using HiScript II First Strand cDNA Synthesis kit (Vazyme, Nanjing, China) according to the manufacturer’s instruction. The mRNA level of IDH1 was determined by quantitative real-time PCR analysis using qPCR SYBR Green Master mix (Vazyme) which was normalized to 18S RNA as the internal control. IDH1 primer: GACTCAGTCGCCCAAGGT (Sense), GCAGCCTCTGCTTCTACC (Antisense); Nrf2 primer: TGGACGGGACTATTGAAGGCTG (Sense), GCCGCCTTTTCAGTAGATGGAGG (Antisense); 18S RNA primer: CTCAACACGGGAAACCTCAC (Sense), CGCTCCACCAACTAAGAACG (Antisense). Each sample was analyzed in triplicate, and the expression levels were normalized using the 2−ΔΔCt -based fold change method.
Preparation of whole-cell lysate
Whole-cell lysate was extracted from cultured JB6 P+ cells in RIPA lysis buffer (Thermo Fisher Scientific, Waltham, MA, USA) containing the protease inhibitor cocktail (Thermo Fisher Scientific). The supernatant collected to measure the protein concentration was detected by BCA assay (CWBIO, CW0014S).
Western blot analysis
Forty micrograms of whole-cell lysate were separated by 10% SDS-PAGE and transferred onto PVDF membranes. Antibodies against IDH1 (1:5,000), ubiquitin (1:2,000) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 1:5,000) were purchased from Abcam (Cambridge, MA, USA). Antibody against β-actin (1:1,000), lactate dehydrogenase (LDH; 1:1,000), glucose transporter 1 (Glut1; 1:1,000), and HIF-1α (1:1,000) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The membranes were incubated with horseradish peroxidase conjugated anti-rabbit or anti-mouse secondary antibodies (1:5,000, Thermo Fisher Scientific). The blots were detected by Biokit Technology Maxilumin™-WB Pico Chemiluminescence Substrate reagent (Baizhi, Bejing, China) and exposed to GeneGnome XRQ chemiluminescence film (Gene Company Limited, Winooski, VT, USA).
Proteasome 20S activity assay
JB6 P+ cells were seeded in a 96-well plate before treated with TPA and/or WA. Proteasome activity was detected using Proteasome 20S Activity Assay Kit (Sigma) according to the manufacturer’s instruction. Proteasome assay loading solution was added to each well, and the plate was incubated for 2 h at 37 °C protecting from light. The fluorescence densities were monitored at λex =490 nm and λem =525 nm. The proteasome activity was calculated according to manufacturer’s instructions (Gene Company Limited).
Immunoprecipitation
For identification of ubiquitinated IDH1 protein, 2 mg of whole-cell lysate were freshly prepared. An aliquot of the supernatant was incubated with anti-isocitrate dehydrogenase antibody (6 µL Abs to 2,000 µg protein in 500 µL cell lysate) overnight at 4 °C with gentle rotation. 25 µL of pre-cooled protein G agarose slurry (Cell Signaling Technology, Danvers, MA, USA) was then added and incubated at 4 °C for 2 hours to capture the immune-complex, which was then centrifuged at 12,000×g at 4 °C for 2 minutes to collect the conjugates. After washing three times, the conjugates were resuspended with 2 times loading buffer, which were then analyzed by western blot analysis. The membranes were hybridized with primary antibodies followed by incubation with appropriate HRP-conjugated secondary antibodies.
IDH1 siRNA and vector transfections
Cells were seeded at 2×105 per well in 6-well tissue culture plates, and transfection was started when cells became 70–80% confluent. For each transfection, 2 µL of the IDH1 siRNA duplex (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was diluted into 100 µL of siRNA transfection medium (Santa Cruz Biotechnology) and labeled as Solution A. Solution B consisted of 2 µL of transfection reagent (Santa Cruz Biotechnology) diluted into 100 µL of siRNA transfection medium. Solution A and B were mixed gently and incubated for 30 min at room temperature. Cells were washed once with 2 mL of siRNA transfection medium. For each transfection, 0.8 mL of siRNA transfection medium was added to each tube containing the solution A/B mixture, mixed and directly added to the washed cells. Six hours later, 2 mL of 1× normal growth medium was added to the cells containing the transfection mixture. The cells were incubated for additional 24 h and subjected to western blot analysis.
For IDH1 overexpression, the cells were washed with PBS and incubated with Opti-MEM medium (Gibco) for 30 min. Five microliter of Lipofectamine 2000 Transfection Reagent (Invitrogen by Thermo Fisher Scientific, Waltham, MA, USA) was diluted with Opti-MEM medium into 125 µL, 2.5 µg of pcDNA3.1 vector or pcDNA3.1-IDH1 plasmid was diluted with Opti-MEM medium into 125 µL. The diluted DNA was added to Lipofectamine 2000 reagent (lipo: DNA ratio =1:1), followed by mixed gently and incubated for 5 min at room temperature. For each transfection, the DNA-lipid complex was added to cells. Four hours later, the Opti-MEM containing the transfection complx was replaced by normal growth medium. After 48 h, cells were collected and assayed.
Alpha-ketoglutarate quantitative assay
Cells treated with TPA and/or WA were collected and resuspended with α-KG Buffer. The supernatant was deproteinized by passing through a 10 kD MWCO spin filter (Thermo Scientific Pierce). Two microliters of the whole-cell lysate filtrate was diluted to 50 µL with α-KG Buffer. Alpha-ketoglutarate concentrations were measured using α-Ketoglutarate Assay Kit (Sigma) according to the manufacturer’s instructions. For each sample, the concentration of α-KG was recorded by spectrophotometry at 570 nm, and calculated by the equation provided by the manufacturer (Gene Company Limited).
Mitochondrial complex I activity assay
The collected cells were lysed by homogenization. The supernatant was collected by centrifugation at 600 g for 5 min at 4 °C. After then, the precipitate was collected by centrifugation at 11,000 g for 10min at 4 °C. Mitochondrial samples were obtained followed by the precipitate was ultrasonically disrupted on ice. Mitochondrial complex I activity was measured using Mitochondrial Respiratory Chain Complex I Activity Assay Kit (Solarbio, Beijing, China) according to the manufacturer’s instruction. The content of complex I was determined at the initiation and 2-min later respectively, which was recorded by spectrophotometry at 340 nm, and calculated by the equation provided by the manufacturer (Gene Company Limited).
LDH activity assay
Cells were resuspended by LDH assay buffer, and then the supernatant was collected. LDH activity was measured using LDH Activity Assay Kit (Sigma) according to the manufacturer’s instructions. The content of LDH was monitored by spectrophotometry at 450 nm, and calculated by the equation provided by the manufacturer (Gene Company Limited).
Mouse vascular endothelial cell growth factor quantitative assay
Cells gathered were resuspended with PBS, and the process that samples frozen at −80 °C for 10 min was repeated three times. The collected supernatant was the sample solution applied to measure the concentration of vascular endothelial growth factor (VEGF). Mouse VEGF concentration was measured using Mouse VEGF ELISA Kit (Yuanmu Biological Technology, Shanghai, China) according to the manufacturer’s instructions (Gene Company Limited).
PHD quantitative assay
Cells gathered were resuspended and frozen at −80 °C for 10 min, which was repeated three times. The collected supernatant was applied to measure the concentration of proline hydroxylase (PHD). PHD concentration was measured using PHD ELISA Kit (Xinyu Biological Technology, Shanghai, China) according to the manufacturer’s instructions, which was recorded by spectrophotometry at 450 nm, and calculated by the equation from manufacturer recommended (Gene Company Limited).
Statistical analysis
One-way analysis of variance (ANOVA) followed by Newman-Keuls post-test was used for multi-group comparisons. All of the experiments have been repeated at least three times. P<0.05 was considered significant.
Results
Downregulation of IDH1 is predominantly due to tumor promoter TPA-induced protein degradation
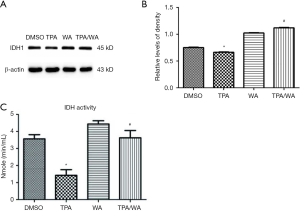
As an important metabolic enzyme, IDH1 reversibly converts isocitrate to α-ketoglutarate in cytosol. Previously we have demonstrated that WA inhibits tumor promoter TPA-induced IDH1 inactivation in JB6 P+ cells (Figure S1). To detect if the expression of IDH1 could be regulated in transcription level, the mRNA levels of IDH1 were detected. Data showed that the IDH1 mRNA levels were no significantly changed after TPA and/or WA treatment (Figure 1A). These results suggest that IDH1 might be regulated at translation or post-translation level but not in transcription level.
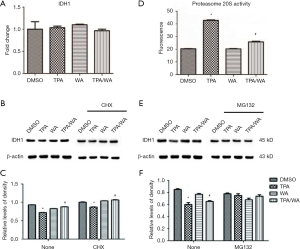
To further investigate how IDH1 was regulated, a protein synthesis inhibitor cycloheximide (CHX) was employed. JB6 P+ cells were pre-treated with CHX for 4 hours and then treated with TPA and/or WA for 24 hours. The effect of WA on IDH1 protein levels was not changed when cells were pretreated with CHX (Figure 1B,C). Therefore, we speculated WA may be able to stabilize IDH1 at the post-translation stage.
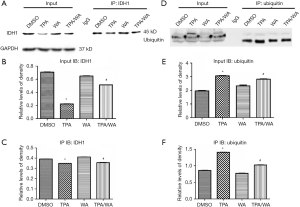
The inhibition of target protein degradation through the ubiquitin-proteasome proteolytic pathway which plays an essential role in the protein metabolism is a recently developed approach to cancer treatment. Proteasome 26S is the most common form of the proteasome, which contains one 20S core particle structure. We first measured the proteasome activity in JB6 P+ cells after treated by TPA and/or WA. TPA-induced significantly increases in proteasome activities (Figure 1D), which contributed to the degradation of protein by activating the proteasome pathway. However, WA attenuated the positive effect of TPA on proteasome (Figure 1D), thereby maintaining protein stability. When the proteasome inhibitor MG132 was applied, the protein levels of IDH1 were restored to normal levels (Figure 1E,F). From the effect of WA/TPA on JB6 P+ cells decreased by MG132 pretreated, we speculated that WA prevented the downregulation and degradation of IDH1 via inhibiting the UPP. For further investigation, we detected the ubiquitination of IDH1. IDH1 proteins were immuno-precipitated, and detected with an anti-ubiquitin antibody. IDH1 expression levels were significantly increased after WA treatment in contrast with that TPA treatment both in the whole-cell lysate and the lysate followed by immunoprecipitation (Figure 2A,B,C). However, increased ubiquitinated IDH1 was observed after TPA treatment before and after immunoprecipitation, indicating that TPA induced the ubiquitination and degradation of IDH1 protein (Figure 2D,E,F). In contrast, WA treatment reduced the ubiquitinated IDH1 and proteasome activity. These results further demonstrate that WA could stabilize IDH1 by inhibiting the UPP.
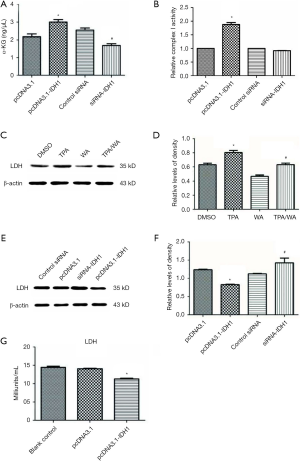
IDH1, as a possible tumor suppressor, inhibits aerobic glycolysis and maintains mitochondrial functions
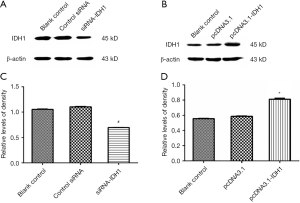
To explore how altered levels of IDH1 affects cellular metabolism, we either overexpressed or knocked down IDH1 in JB6 P+ cells. At first, JB6 P+ cells were transfected with either control siRNA or siRNA to IDH1, and the transfection efficiency was monitored by the IDH1 expression (Figure S2A,B). Similarly, cells were transfected with the pcDNA3.1 or pcDNA3.1-IDH1 vector, and the transfection efficiency was also detected by the IDH1 expression (Figure S2C,D). The concentration of α-KG, the product of IDH1, was detected in these transfected cells. Alpha ketoglutarate levels were significantly increased in pcDNA3.1-IDH1 compared with siRNA-IDH1-transfected cells (Figure 3A). Next, we measured mitochondrial complex I activity, a marker for mitochondrial functions. The complex I activity was significantly increased in pcDNA3.1-IDH1-transfected, but slightly decreased in siRNA-IDH1-transfected cells (Figure 3B). The collected data mirrored that IDH1 functioned in protecting the mitochondria from dysfunction. To observe the impact of IDH1 on glycolysis in carcinogenesis, the expression levels of LDH which is the essential enzyme involved in glycolysis, were detected. WA suppressed the TPA-induced upregulation of LDH expression (Figure 3C,D). WA as an anti-tumor agent might block the tumorigenesis on account of reducing the energy supply provided by glycolysis. Overexpression of IDH1 also inhibited LDH expression, which was reversed by knockdown of IDH1 (Figure 3E,F). In addition, the LDH activity was determined by using the same samples collected from JB6 P+ cells which were transfected with pcDNA3.1-IDH1. IDH1 overexpression reduced the activity of LDH (Figure 3G). These observations indicate that IDH1 might ward off tumorigenesis just as WA, which was mediated by the inhibition on glycolysis.
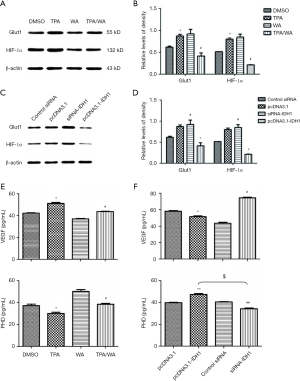
WA suppresses the HIF-1α pathway
Hypoxia inducible factor-1α (HIF-1α) is indispensable in hypoxia, which regulates the biological metabolism of cancer cells to maintain their proliferation. As a cofactor, α-KG produced from IDH1 regulates the activity of PHD, and PHD inhibits HIF-1α activity. Here we measured the expression of HIF-1α targeted genes, including glucose transport protein (Glut) and VEGF. The expression levels of Glut1 and HIF-1α were increased after TPA treatment, which were inhibited by WA (Figure 4A,B). IDH1 overexpression also inhibited the expression of Glut and HIF-1α, but the downregulation of IDH1 enhanced its expression (Figure 4C,D). It was consistent that WA and IDH1 overexpression had the similar impact on the downstream of HIF-1α. Then, the results revealed that WA decreased the levels of VEGF (upper panel), but increased the levels of PHD (lower panel) (Figure 4E). The reduced levels of VEGF by either WA or IDH1 overexpression (Figure 4F) were consistent with the results of Glut1 and HIF-1α, which demonstrated that WA might be an inhibition agent to HIF-1α pathway. The finding that increased level of PHD coincided with the accumulation of α-KG when IDH1 was overexpressed also further supported the connection between PHD and IDH1. Collectively, our results indicate that WA stabilizes IDH1 during tumor promotion, and maintains the activity of PHD which inhibits the activation of HIF-1α, serving as an important mechanism of chemoprevention.
Discussion
IDH1 converts isocitrate to α-KG in the cytoplasm, which activates dioxygenases. Another metabolic enzyme glutamate dehydrogenase (GDH) also converts glutamate to α-KG (24). Our early studies have demonstrated neither TPA nor WA treatment alters GDH activity (16), suggesting that the changes in the α-KG levels mainly come from IDH. Alpha-KG shuttles between the cytosol and mitochondria so that IDH1 is indirectly involved in the tri-carboxylic acid cycle (TCA) (25). Our data have demonstrated that elevated levels of IDH1 increases whereas reduced levels of IDH1 decreases mitochondrial complex I activity, suggesting that IDH1 could maintain the normal mitochondrial functions. Tumorigenesis is often associated with a metabolic shift. Cancer cells predominantly produce energy by a high rate of glycolysis followed by lactic acid fermentation even in the presence of oxygen, known as the “Warburg Effect” (1). Our data show that WA could significantly block the expression of LDH, and overexpression of IDH1 also inhibits both the expression and activity levels of LDH, suggesting a possible mechanism that IDH1 suppresses tumor promotion via inhibiting glycolysis.
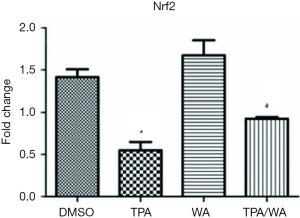
WA is a bioactive compound derived from Withania somnifera, which has been used in clinic for the treatment of nervous and sexual disorders (8). Our preliminary studies have shown that WA inhibited skin tumorigenesis both in vitro and in vivo combined with the overexpression and increased enzyme activity of IDH1 (16,26). Nevertheless, how WA acts as a chemopreventive agent mediated by IDH1 is unclear. It is timely and significant to understand the mechanism of WA in chemoprevention. Our early studies show that WA could block tumor promoter TPA-induced inactivation of IDH1 during tumor promotion (16). As a target gene of nuclear factor erythroid 2-related factor 2 (Nrf2), the promoter of IDH1 includes the antioxidant response element (ARE) which is the binding site of Nrf2 (27,28). We measured the mRNA level of Nrf2 (Figure S3), and results show that TPA treatment suppresses the mRNA levels of Nrf2, which is blocked by WA. However, the mRNA levels of IDH1 are not significantly changed during these treatments (Figure 1A), suggesting the effects on IDH1 are not due to its gene transcription. When applying the protein synthesis inhibitor CHX, the protein levels of IDH1 do not show significant differences between different treatments, suggesting the effects on IDH1 are not due to protein synthesis or the modification of post-translation. Therefore, we focus on whether WA stabilizes IDH1 during post-translation stage. There are two major and fundamentally different mechanisms by which cells degrade proteins, the lysosome and the proteasome (29,30). The UPP is a principal and highly effective protein-degradation pathway in the eukaryotic cells (31), which regulates a wild range of cellular biology including cell growth (32), cell signaling (33), cell immunity (34), cell differentiation (35), and other important cellular physiological processes. The disturbance in the ubiquitination process may lead to tumor promotion (36). WA has showed anti-angiogenesis pharmacological activities through targeting the UPP (37). Therefore, we measure the proteasome activity and in deed, WA suppresses TPA induced proteasome activity. Followed by employing the proteasome inhibitor MG132, WA prevented the downregulation and degradation of IDH1 induced by tumor promoter TPA via inhibiting the UPP. It is further demonstrated by results showing that WA blocks TPA-induced ubiquitination of IDH1, which is consistent with the function of WA as a proteasome inhibitor (38). Since the modification of IDH1 by ubiquitin is of great significance in regulating its degradation and activity, thereby furthermore generates biological effect, what kind of ubiquitinations occurs in IDH1? Which sites of IDH1 are modified by ubiquitin? The studies on the detection of ubiquitination sites are further conducting to explore the detail mechanism.
As a downstream molecular of α-KG, PHD inhibits the HIF-1α-induced signal pathway involved in tumorigenesis (39,40). Our data are showing that PHD activity is induced by WA treatment, as well IDH1 overexpression, establishing a link WA to HIF-1α. It is further supported by the data that WA blocks TPA-induced activation of Glut1 and VEGF, both the target molecules of HIF-1α and contributing to tumorigenesis (41,42).
Conclusions
Taken together, our present study has demonstrated a novel mechanism of how WA, as a potential chemopreventive agent, inhibits tumor promotion, starting from stabilizing IDH1 to inactivating HIF-1α signaling. These results uncover the mechanism of tumor suppressor gene IDH1 as a promising molecular target for chemoprevention, and WA might provide a guide for the discovery of chemopreventive chemicals.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.09.57). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The research was conducted at the molecular and cellular levels, and didn’t employ the animal model. The ethical statement was not required.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Warburg O. On the origin of cancer cells. Science 1956;123:309-14. [Crossref] [PubMed]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009;324:1029-33. [Crossref] [PubMed]
- Rohle D, Popovici-Muller J, Palaskas N, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 2013;340:626-30. [Crossref] [PubMed]
- Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010;18:553-67. [Crossref] [PubMed]
- Yang Z, Jiang B, Wang Y, et al. 2-HG Inhibits Necroptosis by Stimulating DNMT1-Dependent Hypermethylation of the RIP3 Promoter. Cell Rep 2017;19:1846-57. [Crossref] [PubMed]
- Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009;462:739-44. [Crossref] [PubMed]
- Laba P, Wang J, Zhang J. Low level of isocitrate dehydrogenase 1 predicts unfavorable postoperative outcomes in patients with clear cell renal cell carcinoma. BMC Cancer 2018;18:852. [Crossref] [PubMed]
- Chowdhury K, Neogy RK. Mode of action of Withaferin A and Withanolide D. Biochem Pharmacol 1975;24:919-20. [Crossref] [PubMed]
- Liu X, Chen L, Liang T, et al. Withaferin A induces mitochondrial-dependent apoptosis in non-small cell lung cancer cells via generation of reactive oxygen species. J BUON 2017;22:244-50. [PubMed]
- Zhang HL, Zhang H. Withaferin-A Induces Apoptosis in Osteosarcoma U2OS Cell Line via Generation of ROS and Disruption of Mitochondrial Membrane Potential. Pharmacogn Mag 2017;13:523-27. [Crossref] [PubMed]
- Ndlovu MN, Van Lint C, Van Wesemael K, et al. Hyperactivated NF-{kappa}B and AP-1 transcription factors promote highly accessible chromatin and constitutive transcription across the interleukin-6 gene promoter in metastatic breast cancer cells. Mol Cell Biol 2009;29:5488-504. [Crossref] [PubMed]
- Srinivasan S, Ranga RS, Burikhanov R, et al. Par-4-dependent apoptosis by the dietary compounwithaferin A in prostate cancer cells. Cancer Res 2007;67:246-53. [Crossref] [PubMed]
- Koduru S, Kumar R, Srinivasan S, et al. Notch-1 inhibition by Withaferin-A: a therapeutic target against colon carcinogenesis. Mol Cancer Ther 2010;9:202-10. [Crossref] [PubMed]
- Yu Y, Hamza A, Zhang T, et al. Withaferin A targets heat shock protein 90 in pancreatic cancercells. Biochem Pharmacol 2010;79:542-51. [Crossref] [PubMed]
- Walker K, Padhiar M. AACR-NCI-EORTC--21st International Symposium. Molecular targets and cancer therapeutics--Part 1. IDrugs 2010;13:7-9. [PubMed]
- Li W, Zhao Y. Withaferin A suppresses tumor promoter 12-O-tetradecanoylphorbol 13-acetate-induced decreases in isocitrate dehydrogenase 1 activity and mitochondrial function in skin epidermal JB6 cells. Cancer Sci 2013;104:143-8. [Crossref] [PubMed]
- Khedgikar V, Kushwaha P, Gautam J, et al. Withaferin A: a proteasomal inhibitor promotes healing after injury and exerts anabolic effect on osteoporotic bone. Cell Death Dis 2013;4:e778. [Crossref] [PubMed]
- Zhang X, Timmermann B, Samadi AK, et al. Withaferin a induces proteasome-dependent degradation of breast cancer susceptibility gene 1 and heat shock factor 1 proteins in breast cancer cells. ISRN Biochem 2012;2012:707586. [Crossref] [PubMed]
- Liu J, Shaik S, Dai X, et al. Targeting the ubiquitin pathway for cancer treatment. Biochim Biophys Acta 2015;1855:50-60. [PubMed]
- Rossi M, Aqeilan RI, Neale M, et al. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc Natl Acad Sci U S A 2006;103:12753-8. [Crossref] [PubMed]
- Wang X, Trotman LC, Koppie T, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 2007;128:129-39. [Crossref] [PubMed]
- Grover A, Shandilya A, Bisaria VS, et al. Probing the anticancer mechanism of prospective herbal drug Withaferin A on mammals: a case study on human and bovine proteasomes. BMC Genomics 2010;11:S15. [Crossref] [PubMed]
- Yang H, Shi G, Dou QP. The tumor proteasome is a primary target for the natural anticancer compound Withaferin A isolated from "Indian winter cherry". Mol Pharmacol 2007;71:426-37. [Crossref] [PubMed]
- Lu Z, Scott I, Webster BR, et al. The emerging characterization of lysine residue deacetylation on the modulation of mitochondrial function and cardiovascular biology. Circ Res 2009;105:830-41. [Crossref] [PubMed]
- Frezza C, Tennant DA, Gottlieb E. IDH1 mutations in gliomas: when an enzyme loses its grip. Cancer Cell 2010;17:7-9. [Crossref] [PubMed]
- Li W, Zhang C, Du H, et al. Withaferin A suppresses the up-regulation of acetyl-coA carboxylase 1 and skin tumor formation in a skin carcinogenesis mouse model. Mol Carcinog 2016;55:1739-46. [Crossref] [PubMed]
- Xu C, Huang MT, Shen G, et al. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res 2006;66:8293-6. [Crossref] [PubMed]
- Chen C, Kong AN. Dietary chemopreventive compounds and ARE/EpRE signaling. Free Radic Biol Med 2004;36:1505-16. [Crossref] [PubMed]
- Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self-digestion. Nature 2008;451:1069-75. [Crossref] [PubMed]
- Reinstein E, Ciechanover A. Narrative review: protein degradation and human diseases: the ubiquitin connection. Ann Intern Med 2006;145:676-84. [Crossref] [PubMed]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem 1998;67:425-79. [Crossref] [PubMed]
- Benanti JA. Coordination of cell growth and division by the ubiquitin-proteasome system. Semin Cell Dev Biol 2012;23:492-98. [Crossref] [PubMed]
- Willis MS, Townley-Tilson WH, Kang EY, et al. Sent to destroy: the ubiquitin proteasome system regulates cell signaling and protein quality control in cardiovascular development and disease. Circ Res 2010;106:463-78. [Crossref] [PubMed]
- Wang J, Maldonado MA. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell Mol Immunol 2006;3:255-61. [PubMed]
- Naujokat C, Saric T. Concise review: role and function of the ubiquitin-proteasome system in mammalian stem and progenitor cells. Stem Cells 2007;25:2408-18. [Crossref] [PubMed]
- Chen X, Shen J, Li X, et al. Rlim, an E3 ubiquitin ligase, influences the stability of Stathmin protein in human osteosarcoma cells. Cell Signal 2014;26:1532-38. [Crossref] [PubMed]
- Mohan R, Hammers HJ, Bargagna-Mohan P, et al. Withaferin A is a potent inhibitor of angiogenesis. Angiogenesis 2004;7:115-22. [Crossref] [PubMed]
- Yang H, Wang Y, Cheryan VT, et al. Withaferin A inhibits the proteasome activity in mesothelioma in vitro and in vivo. PLoS One 2012;7:e41214. [Crossref] [PubMed]
- Mimeault M, Batra SK. Hypoxia-inducing factors as master regulators of stemness properties and altered metabolism of cancer- and metastasis-initiating cells. J Cell Mol Med 2013;17:30-54. [Crossref] [PubMed]
- Partch CL, Gardner KH. Coactivators necessary for transcriptional output of the hypoxia inducible factor, HIF, are directly recruited by ARNT PAS-B. Proc Natl Acad Sci U S A 2011;108:7739-44. [Crossref] [PubMed]
- Armitage EG, Kotze HL, Allwood JW, et al. Metabolic profiling reveals potential metabolic markers associated with Hypoxia Inducible Factor-mediated signalling in hypoxic cancer cells. Sci Rep 2015;5:15649. [Crossref] [PubMed]
- Lee SY, Kim HJ, Oh SC, et al. Genipin inhibits the invasion and migration of colon cancer cells by the suppression of HIF-1alpha accumulation and VEGF expression. Food Chem Toxicol 2018;116:70-6. [Crossref] [PubMed]


