
Up-regulated GGA3 promotes non-small cell lung cancer proliferation by regulating TrkA receptor
Introduction
Speaking of malignant tumors worldwide, lung cancer must be mentioned, with high morbidity and mortality. Nearly 90% of lung cancer patients are diagnosed with non-small cell lung cancer (NSCLC) (1). Although many treatment including surgery, chemotherapy, radiotherapy, and molecular target therapy are available for NSCLC, the survival rate is still very low for the high recurrence rate, toxic side effects and poor efficacy (2). Therefore, further understanding of the pathogenesis of NSCLC is helpful to search for the effective therapeutic target for NSCLC.
Golgi-localized gamma-ear including Arf-binding protein 3 (GGA3) was monolecular adaptor involved in the trans-Golgi network (TGN) and endosomes that function in the intracellular transport of receptors (3). GGAs perform as adaptors for the accurate sorting of vesicle between the TGN and endosomes, which ensure the efficient transport of various types of enzymes and accurate transport or recovery of membrane proteins (4). As a member of GGAs family, GGA3 has been found to be related to cellular events such as cell survival, cell migration and cell apoptosis through different molecular mechanism in several cancers. Ratcliffe et al. reported that GGA3 could modulate SNX17 endosomal localization and interact with Arf6 transferred integrins of the degradation pathway, further contribute to sustain cell migration (5). Li et al. reported that GGA3 regulated the endocytic cycle of TrkA and promoted the continuous phosphorylation of Akt and cell survival (6). Inhibition of endosomal regulators GGA3 may perturb the c-Met endosomal signaling for hepatocellular carcinoma cell migration (7). However, the direct relationship between GGA3 and NSCLC has not been reported yet.
In this research, we intended to investigate the influence of GGA3 on NSCLC cell proliferation, apoptosis, invasion and migration. Furthermore, the underlying mechanism of GGA3 in NSCLC progression was explored either.
Methods
Data collection
Data from The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/), which includes 535 NSCLC tissues and 59 normal tissues, were used to analyze expression of GGA3 and TrkA, and correlation between GGA3 and TrkA.
Cell culture
The Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China) provided the Human NSCLC cell A549, H157, H2087 and human bronchial epithelial cell BEAS-2B. Cells were maintained with Roswell Park Memorial Institute medium (RPMI-1640; HYCLONE, USA) at the condition of 37 °C, 5% CO2. The cell culture medium included 10% fetal bovine serum (FBS), 0.1 mg/mL streptomycin (Sigma, USA), and 100 U/mL penicillin (Sigma, USA). As the cells growing to the logarithmic growth phase, cleaned the cells and digested by 0.25% trypsin (Solarbio, Beijing) until the cells becoming round, re-added the cell culture medium to stop the digestion. Prepared of cell suspension and seeded to 6-well plates for the next assays.
Cell transfection
After the A549 cells density reached about 80%, cells were processed for transfection. Cells maintained until mid-log phase into 6-well plates (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), and fresh cell culture medium was infused two hours prior to transfection. PcDNA3.1-GGA3 group was transfected with pcDNA3.1-GGA3 (synthesized by GeneChem, Shanghai, China), and the control group was transfected with pcDNA3.1 empty vector by Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in line with the supplier’s instruction. Ten µL Lipofectamine 2000 was dissolved in 250 µL of serum-free antibiotic-free medium, mixed gently and allowed to stand for 5 minutes at room temperature. Then 5 µL plasmid was dissolved in 250 µL serum-free antibiotic-free medium. Mixed the liposome and plasmid mixture gently within 30 min. Then discarded the medium from the 6-well plate and washed the cells twice with PBS. Added 500 µL of liposome and plasmid mixture to each well of cells, gently mixed and incubated in incubator for 6 h, replaced the mixture with cell culture medium. After 24 h incubation, we can observe the expression of plasmids or proceed with follow-up experiments.
Reverse transcription-polymerase chain reaction (RT-PCR)
Isolated total RNA through TRIzol reagent (CWBio, Beijing, China). To detect the mRNA expression of GGA3, firstly, total RNA was reverse-transcribed into cDNA through the PrimeScript RT Reagent Kit (TaKaRa, Japan) in lines with the supplier’s instruction, then using 1 µg cDNA to proceed PCR by TaqMan® Universal Master Mix (Life technology, Carlsbad, CA, USA) according to the supplier’s instruction. The procedure was consisted of 40 amplification cycles (95 °C for 30 sec, 60 °C for 45 sec) and 72 °C for 30 min. Primers (Sangon Biotech Co., Ltd., Shanghai, China) used for GGA3 were presented as follows: forward, 5'-CAGGGTGTCTGAGAAAGTGAAG-3' and reverse, 5'-CATGTGGTAGGCGTCTTTGA-3'. β-actin was as an internal control, β-actin forward: 5'-CCCGAGCCGTGTTTCCT-3', reverse: 5'-GTCCCAGTTGGTGACGATGC-3'. Three replicate wells were set up for each sample, and the results were analyzed by 2−△△Ct method.
Western blot
Transfected with plasmid for 48 h, placed the six-well plate on ice and treated with radio immunoprecipitation assay (RIPA; Cwebio, Beijing) lysis buffer including protease inhibitor (Cwebio, Beijing). Then extracted the protein and measured the concentration by BCA kit (Cwebio, Beijing). Extracted proteins (20 µg) mixed with loading buffer (5×; Thermo, USA) were denatured with boiling water for 5 min. The mixture of denatured protein was separated by polyacrylamide gel (Biorad, USA) and transferred to polyvinylidene fluoride (PVDF; Thermo, USA) membrane. The membrane was sealed with 5% skim milk for 1 h and incubated with primary antibodies of relevant proteins overnight at 4 °C. Washed the membrane for 5 min with three times and incubated with secondary antibody for 1 h. Signal was enhanced by ECL reagent (PTG, USA), and quantitated by QUANTITY ONE after the final washing, used GAPDH as an internal control. The antibodies used were shown as follows: Bax (1:1,000, PTG, USA), Bcl-2 (1:1,000, PTG, USA), TrkA (1:1,000, Abcam, USA), Active-caspase 3 (1:1,000, PTG, USA), AKT (1:1,000, CST, USA), p-AKT (1:1,000, CST, USA), ERK (1:1,000, CST, USA), p-ERK (1:1,000, CST, USA), GAPDH (1:5,000, PTG, USA); polyclonal antibody HRP-conjugated goat anti- rabbit (1:5,000, PTG, USA).
Cell proliferation assay
Cells transfected for 24 h were digested for count number, and prepared for cell suspension. Seeded 100 µL cell suspension in 96-well plate with the density of 1×103 cells/well. Cells were incubated in carbon dioxide incubator, and cell viability was detected every 24 h. Before the optical density (OD) 450 nm value was detected, 10 µL of CCK8 reagent (Solarbio, Beijing, China) was placed into the cell supernatant, and cultured for 1.5 h at 37 °C. Plotted the proliferation curve. The result was expressed as the mean value ± standard deviations (SD).
Transwell assays
A 24-well transwell plate was used to perform the invasion assay. One hundred µL Matrigel (BD, USA), which diluted at 1:6 with serum-free medium overnight, was placed to the transwell chamber, and cultured for 4–6 h in carbon dioxide incubator at 37 °C. Then the upper medium was suck up, added 500 µL DMEM medium without FBS to the lower chambers to hydrate basement membrane. After the cells were transfected for 24 h, 100 µL cell suspension (1×105 cells) was seeded to the upper chambers, 500 µL DMEM medium with 10% FBS was filled in the lower chambers, incubated at 37 °C for overnight. Removed the residual cells in upper chambers by cotton-tipped swab. The cells that invaded were fixed with 4% paraformaldehyde for 30 min, and stained with 0.1% crystal violet for 20 min. Washed with PBS, counted the invading cells randomly and statistical.
The migration experiment process is similar to the invasion assay except without pre-coated with Matrigel.
Flow cytometry analysis
Transfected for 24 h, changed to serum-free medium. Digested cells by trypsin get rid of EDTA, collected for centrifugation. Cells were resuscitated in pre-cooled PBS, centrifuged again and aspirated the supernatant. Re-suspending the cells with 1× binding buffer, with density of 1×106–5×106/mL. Took 100 µL cell suspension and 5 µL Annexin V/FITC (4A Bio, Beijing) into 5 mL flow tube for mixing incubation at room temperature for 5 min behind the scenes. Added 10 µL PI dye and 400 µL PBS for detection. The signals were detected by FlowJo software.
Statistical analysis
Using SPSS 18.0 statistical analysis software to analyze the experimental data. Measurement data showed as mean ± standard (mean ± SD), difference between two groups was calculated through t-test. Comparison among multiple groups was analyzed by ANOVA following post hoc test of Dunnett. P<0.05 was considered as statistically significant.
Results
GGA3 was high expressed in NSCLC tissues and cell lines
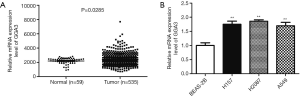
To explore the function of GGA3 in NSCLC, TCGA dataset including 535 NSCLC tissues and 59 normal tissues were employed to detect the expression of GGA3 in NSCLC tissues. As presented in Figure 1A, GGA3 was significantly up-regulated in NSCLC tissues compared with the normal, P=0.0285. Subsequently, we adopted qRT-PCR to analyze the relative expression of GGA3 in several NSCLC cell lines including H157, H2087, and A549. Through the qRT-PCR result, we found that GGA3 was highly expressed in NSCLC cell lines compared to normal human bronchial epithelial cell BEAS-2B, suggesting that GGA3 played a vital role in NSCLC (P<0.05, Figure 1).
Expression of pcDNA3.1-GGA3 in A549 cells
To observe the action of GGA3 in NSCLC cells proliferation, A549 cells were employed and GGA3 overexpressed plasmid was constructed. Firstly, we tested the mRNA and protein expression of GGA3. The result showed that mRNA and protein expression of GGA3 in pcDNA3.1-GGA3 group were increased in contrast with sham group (P<0.05, Figure 2A,B). This finding showed that the pcDNA3.1-GGA3 plasmid could successfully elevate the expression of GGA3 in NSCLC A549 cells.

GGA3 promoted the proliferation of A549 cells
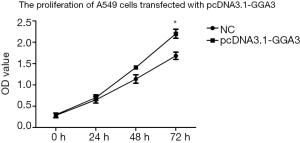
To investigate the function of GGA3 on cell multiplication, we measured the proliferation of NSCLC cell A549 in pcDNA3.1-GGA3 group and the control group, respectively. As shown in Figure 3, the OD value of A549 cells was significantly increased at 72 h in the CCK8 assay after overexpression of GGA3 (P<0.05). This finding revealed that the proliferation of A549 cells was increased after overexpression of GGA3, indicating that GGA3 was capable to promote NSCLC cells growth.
GGA3 promoted the aggressiveness of NSCLC cell A549
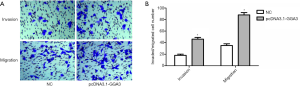
Given that GGA3 promoted the multiplication of NSCLC cell A549, we then investigated the roles of GGA3 in cell migration and invasion in A549. The finding showed that the number of invaded cells in pcDNA3.1-GGA3 group (46±3) was obviously increased compared to sham group (18±2) (P<0.05, Figure 4A,B). The migrated cells in pcDNA3.1-GGA3 group (88±4) was also obviously enhanced compared to control group (35±3) (P<0.05, Figure 4A,B). These findings showed that GGA3 could significantly induce cell migration and invasion abilities in NSCLC cell A549.
GGA3 inhibited the apoptosis of NSCLC cell A549
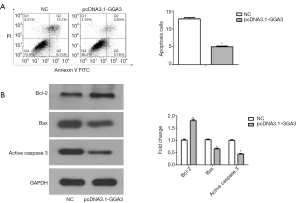
Annexin V staining of A549 cells (annexin-V positive and FITC positive) explored apoptosis of a proportion of cells among the adherent cells at 24 h after transfection. Flow cytometry analysis showed that apoptotic cells were especially decreased in pcDNA3.1-GGA3 group (5.02%) in contrast with the control (12.88%) (Figure 5A). These findings suggested that overexpressed GGA3 could reduce cell apoptosis. To further validate GGA3 reduced apoptosis, the pro-apoptotic protein Active caspase-3 and Bax, as well as the anti-apoptotic protein Bcl-2 were detected in A549 cells. As presented in Figure 5B, the expression of Bax and Active caspase-3 were decreased, whereas the expression level of Bcl-2 was increased in A549 cells in pcDNA3.1-GGA3 group in contrast with the sham group (P<0.05). Altogether, these findings demonstrated that GGA3 can suppress apoptosis in A549 NSCLC cells.
GGA3 activated AKT/ERK signaling pathway in NSCLC cell A549
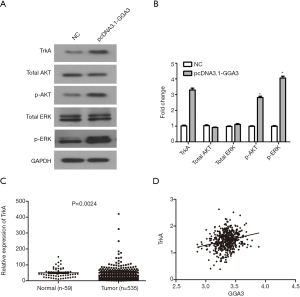
Previous research has shown that apoptosis was positively correlated with AKT/ERK signaling pathway. To explore whether GGA3-mediated apoptosis in A549 cells through the specific signaling pathway, we detected the signaling proteins related to the activate process, including AKT, p-AKT, ERK, p-ERK. We found that the expression of p-AKT and p-ERK in pcDNA3.1-GGA3 group were higher than control group (Figure 6A,B). Furthermore, the expression of TrkA was obviously increased compared with control group (Figure 6A,B). All findings suggested that the cell effects induced by GGA3 might be mediated by overexpression of TrkA. In addition, according to TCGA data analysis, TrkA was found to be highly expressed in NSCLC tissues (Figure 6C). And the correlation analysis results showed that GGA3 expression and TrkA expression had a certain positive relationship in NSCLC (Figure 6D). In a word, GGA3 significantly activated AKT/ERK signaling pathway in NSCLC cell A549 through TrkA receptor.
Discussion
Our results showed that GGA3 was highly expressed in NSCLC tissues and cell lines. Overexpression of GGA3 could prompt the growth, aggressiveness, whereas inhibit the apoptosis of A549 cells. Finally, it has been proved that GGA3 promoted NSCLC cells cancer-related behaviors partly through regulating TrkA-AKT/ERK signaling pathway.
GGA3 is one member of three GGA subtypes (GGA1, GGA2, and GGA3) in humans with similar transportation functions (8). All three GGAs are made up of four segments: a VHS domain that binds to the acidic dileucine sorting signals (DXXLL, X = any amino acid) exists in the cytoplasmic tail of various transmembrane proteins, for example mannose-6-phosphate receptor (MPR); a GAT domain that coaction with the Arfs and ubiquitin GTP-bounding forms; an unstructured hinge region that binds to clathrin; and a GAE domain, which related to numerous accessory factors (9). The VHS-DXXLL signal interactions could specifically dictate cargo proteins into the Golgi/TGN-to-endosome pathway (10). Among of which, the TGN compartment is famous for the “sorting center” where newly synthesized proteins are classified to their final cellular destinations such as lysosomes, endosomes and the plasma membrane (11). GGA protein is highly conserved and can recognize sorting signals of acidic di-L signal and ubiqutin signal, binds to clathrin or ARF protein, and participates in vesicle formation and fusion by binding to multiple proteins. Precise and controlled transport of vesicles in the vesicle system ensures important functions such as endocytosis, exocytosis, efficient transport of various enzymes in lysosomes, transport and recovery of membrane organelles of various organelles (12,13). Deletion or mutation of the GGA proteins results in disruption of vesicular trafficking between TGN and endosomes (5). Thus GGA proteins are very important for cellular functions especially in cancer cell. It has been reported that GGA3 was related to cell migration, cell proliferation and cell apoptosis in various cells sometimes in carcinoma cells. So in this report, we verified that GGA3 was overexpressed in NSCLC tissues and cell lines. Subsequently, pcDNA3.1-GGA3 was applied to regulate the expression of GGA3. As shown in Figure 2, the expression of GGA3 at mRNA and protein levels was obviously up-regulated after transfected with pcDNA3.1-GGA3 in NSCLC cell A549 in contrast with control group.
Tyrosine kinase receptor A (TrkA), a high-affinity receptor of NGF function as a vital character in developmental neurobiology, is desorbed by proliferating keratinocytes and induces multiplication (14). Once internalized into early endosomes, TrkA could initiate receptor phosphorylated and activate downstream signals transduction cascades, including the phosphatidylinositol 3-kinase (PI3K)/AKT, phospholipase Cγ/protein kinase C (PKC) and Ras/mitogen-activated protein kinase (MAPK) signaling pathways (15). Then TrkA could either be recalled back to the plasma membrane (PM), for functional resensitization and prolongation of cellular surface–specific signaling process, or it can be concentrated into lysosomes for degradation, resulting in extended attenuation of cellular signaling. The separation of TrkA into different pathways has a vital influence on biological functions regulated by TrkA, and its transformation might result in various cellular function variation (15-19). It has been reported that GGA3 directly bound to TrkA internal DXXLL motifs through Arf6 to mediate TrkA recycling, and silenced GGA3 could attenuate TrkA-dependent AKT signaling and cell survival (6). Consistent with this report, we also found that the expression of TrkA was increased in the up-regulation of GGA3 in NSCLC cell A549. Furthermore, TrkA has been reported to prompt cancer cell survival, cell proliferation and cell invasion and so on (14,20-22). Consolidated with these results, in this report the proliferation, migration and invasion of NSCLC A549 cells were prompted, but the apoptosis of A549 cells was inhibited.
In conclusion, our findings suggested that GGA3 could prompt NSCLC cells A549 proliferation, invasion and migration, and inhibit cell apoptosis partly through mediating TrkA-AKT/ERK signaling pathway. This is the first report that highlights the correlation between GGA3 and NSCLC tumorigenesis to our knowledge, which might provide a new potential target for diagnosis and therapy of NSCLC.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.10.17). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional ethical approval and individual informed consent were waived due to the nature of the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wrona A, Jassem J. The new TNM classification in lung cancer. Pneumonol Alergol Pol 2010;78:407-17. [PubMed]
- Nanavaty P, Alvarez MS, Alberts WM. Lung cancer screening: advantages, controversies, and applications. Cancer Control 2014;21:9-14. [Crossref] [PubMed]
- von Einem B, Wahler A, Schips T, et al. The Golgi-Localized gamma-Ear-Containing ARF-Binding (GGA) Proteins Alter Amyloid-beta Precursor Protein (APP) Processing through Interaction of Their GAE Domain with the Beta-Site APP Cleaving Enzyme 1 (BACE1). PLoS One 2015;10:e0129047. [Crossref] [PubMed]
- Doray B, Misra S, Qian Y, et al. Do GGA adaptors bind internal DXXLL motifs? Traffic 2012;13:1315-25. [Crossref] [PubMed]
- Ratcliffe CD, Sahgal P, Parachoniak CA, et al. Regulation of Cell Migration and beta1 Integrin Trafficking by the Endosomal Adaptor GGA3. Traffic 2016;17:670-88. [Crossref] [PubMed]
- Li X, Lavigne P, Lavoie C. GGA3 mediates TrkA endocytic recycling to promote sustained Akt phosphorylation and cell survival. Mol Biol Cell 2015;26:4412-26. [Crossref] [PubMed]
- Hu CT, Wu JR, Cheng CC, et al. The Therapeutic Targeting of HGF/c-Met Signaling in Hepatocellular Carcinoma: Alternative Approaches. Cancers (Basel) 2017;9. [PubMed]
- Santonico E, Mattioni A, Panni S, et al. RNF11 is a GGA protein cargo and acts as a molecular adaptor for GGA3 ubiquitination mediated by Itch. Oncogene 2015;34:3377-90. [Crossref] [PubMed]
- Wakasugi M, Waguri S, Kametaka S, et al. Predominant expression of the short form of GGA3 in human cell lines and tissues. Biochem Biophys Res Commun 2003;306:687-92. [Crossref] [PubMed]
- Kametaka S, Mattera R, Bonifacino JS. Epidermal growth factor-dependent phosphorylation of the GGA3 adaptor protein regulates its recruitment to membranes. Mol Cell Biol 2005;25:7988-8000. [Crossref] [PubMed]
- Kawasaki M, Shiba T, Shiba Y, et al. Molecular mechanism of ubiquitin recognition by GGA3 GAT domain. Genes Cells 2005;10:639-54. [Crossref] [PubMed]
- Tesco G, Koh YH, Kang EL, et al. Depletion of GGA3 stabilizes BACE and enhances beta-secretase activity. Neuron 2007;54:721-37. [Crossref] [PubMed]
- Walker KR, Modgil A, Albrecht D, et al. Genetic Deletion of the Clathrin Adaptor GGA3 Reduces Anxiety and Alters GABAergic Transmission. PLoS One 2016;11:e0155799. [Crossref] [PubMed]
- Aubert L, Guilbert M, Corbet C, et al. NGF-induced TrkA/CD44 association is involved in tumor aggressiveness and resistance to lestaurtinib. Oncotarget 2015;6:9807-19. [Crossref] [PubMed]
- Marlin MC, Li G. Biogenesis and function of the NGF/TrkA signaling endosome. Int Rev Cell Mol Biol 2015;314:239-57. [Crossref] [PubMed]
- Calvo L, Anta B, Lopez-Benito S, et al. Bex3 Dimerization Regulates NGF-Dependent Neuronal Survival and Differentiation by Enhancing trkA Gene Transcription. J Neurosci 2015;35:7190-202. [Crossref] [PubMed]
- Fujimura M, Usuki F. Methylmercury causes neuronal cell death through the suppression of the TrkA pathway: in vitro and in vivo effects of TrkA pathway activators. Toxicol Appl Pharmacol 2015;282:259-66. [Crossref] [PubMed]
- Jung EJ, Park HC, Chung KH, et al. Proteomic analysis of SP600125-controlled TrkA-dependent targets in SK-N-MC neuroblastoma cells: inhibition of TrkA activity by SP600125. Proteomics 2014;14:202-15. [Crossref] [PubMed]
- Prencipe G, Minnone G, Strippoli R, et al. Nerve growth factor downregulates inflammatory response in human monocytes through TrkA. J Immunol 2014;192:3345-54. [Crossref] [PubMed]
- Shi W, George SK, George B, et al. TrkA is a binding partner of NPM-ALK that promotes the survival of ALK (+) T-cell lymphoma. Mol Oncol 2017;11:1189-207. [Crossref] [PubMed]
- Jung EJ, Chung KH, Bae D-W, et al. Proteomic analysis of novel targets associated with the enhancement of TrkA-induced SK-N-MC cancer cell death caused by NGF. Exp Mol Med 2016;48:e235. [Crossref] [PubMed]
- Pasini L, Re A, Tebaldi T, et al. TrkA is amplified in malignant melanoma patients and induces an anti-proliferative response in cell lines. BMC Cancer 2015;15:777. [Crossref] [PubMed]


