
A comparison of MASS-PCR and ARMS-PCR for the detection of lung cancer gene mutation
Introduction
Lung cancer is the most common cause of cancer-related death around the world. Non-small cell lung cancer (NSCLC) accounts for approximately 85% of all lung cancer patients (1). The prognosis of lung cancer is poor, with a 5-year survival rate of 4–17% depending on stage and regional differences (2). In recent decades, much progress has been made in comprehensive genomic profiling of lung cancer and divide the lung cancer into several molecular subtypes according to the oncogenic driver mutations identified in patients, thus contributing to the personalized therapy, especially targeted therapy. At present, the targeted therapies mainly targeted at the common mutations in lung cancer, including EGFR, BRAF, HER2, KRAS, MET, ALK, ROS1, RET, NTRK (3,4). Clinical practices have confirmed the success of targeted therapies in several molecular defined lung cancer subtypes (5). Whereas targeted therapy needs exact mutation detection.
In the past decades, Sanger sequencing was the most commonly used assay for mutation detection. At present, Sanger sequencing remains the gold standard in clinical practice and may detect unknown mutations (6,7). However, it is time-consuming and may delay the clinical-decision making (8). Besides, it has low sensitivity because the mutant tumor cells are required to more than 20% of all detected tumor cells, possibly leading to the false-negative result (9). Currently, many detection assays with higher sensitivity have been developed for mutation detection. Amplification refractory mutation system PCR (ARMS-PCR) has been approved by the China Food and Drug Administration (CFDA) and become a widely used method in clinic practice. ARMS-PCR is more sensitive than Sanger sequencing with its detection limit of about 1% (8). However, because the fluorescent signals are determined through calculating the value of ΔCt, sometimes it is highly subjective and difficult to make the determination of the conclusive results. Mutation-selected amplification specific system PCR (MASS-PCR) is a novel quantitative PCR (qPCR)-based assay to detect mutations. As only mutant gene emits a fluorescence peak in the cyclic amplification, it solved the defect of ARMS-PCR (10). In this study, we compared the accuracy and sensitivity of ARMS-PCR and MASS-PCR and evaluated the clinical feasibility of MASS-PCR, purposed to supply an alternative possibility to be more effective and exact in mutation detection.
Methods
Samples collection and processing
A total of 293 formalin-fixed paraffin-embedded (FFPE) tissues were collected from 293 lung cancer patients in Sun Yat-sen University Cancer Center and Foshan First People’s Hospital from 2017 to 2018. The study was approved by the Institutional Review Board (IRB), Sun Yat-sen University Cancer Center and Foshan First People’s Hospital. Written informed consent was obtained from all participants. Genomic DNA was extracted from FFPE tissue according to the manufacturer’s instruction, and then, the genomic DNA was used to determine the genotype in the following methods.
MASS-PCR assay
MASS-PCR is a qPCR-based assay with specific primers and probes that target at the common mutations of EGFR, KRAS, BRAF, HER2, MET exon 14 skipping mutation, ALK fusion, and ROS1 fusion genes. MASS-PCR was performed in a StepOne™ PCR System (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instruction. The one-step method was applied in all gene mutations detection, including ALK fusion and ROS1 fusion genes. The kit was bought from the Shanghai Tongshu Biotechnology Co., Ltd.
ARMS-PCR assay
The isolated DNA samples were re-tested with ARMS-PCR. ARMS-PCR was used to detect the same mutation as MASS-PCR assay. For the detection of ALK fusion and ROS1 fusion genes, the two-step method was needed. Briefly, RNA was extracted from FFPE tissue and produced into complementary DNA (cDNA) according to the manufacturer’s instruction. cDNA was then used to determine the genotype. The kit was brought from the Beijing ACCB Biotechnology Co., Ltd.
Sanger sequencing
Sanger sequencing was performed to verify the genotype. DNA was used as a template to amplify the involved genes. PCR products were then sequenced using Sanger method.
Statistical analysis
The statistical analysis and graphic generation were performed in SPSS 19.0 and MedCalc 19.0. The concordance rate of MASS-PCR, ARMS-PCR, and Sanger sequencing in identifying mutation status was analyzed by kappa test. P<0.05 was considered to indicate statistical significance.
Results
Clinical information on lung cancer patients
This study recruited 293 patients with lung cancer, in which there were 226 males (77.1%) and 67 females (22.9%) (Table 1). The age of patients ranged from 29 to 86 years old with the median age of 64-year-old. All the detected samples were originated from FFPE tissues. Most patients in our study were diagnosed as NSCLC accounting for 95.2%, in which, squamous cell cancer accounted for 29.4%, and adenocarcinoma accounted for 60.4%. Small cell lung cancer (SCLC) only accounted for 2.0%.
Table 1
| Variables | Number (n=293) | Proportion, % |
|---|---|---|
| Gender | ||
| Male | 226 | 77.1 |
| Female | 67 | 22.9 |
| Age (range, years) | 29–86 | |
| Histological types | ||
| NSCLC | 279 | 95.2 |
| Squamous cell cancer | 86 | 29.4 |
| Adenocarcinoma | 177 | 60.4 |
| Large cell carcinoma | 12 | 4.1 |
| Adenosquamous carcinoma | 1 | 0.3 |
| SCLC | 6 | 2.0 |
| Undefined | 8 | 2.7 |
NSCLC, non-small cell lung cancer; SCLC, small cell lung cancer.
Comparison between MASS-PCR and ARMS-PCR
Here, we conducted the gene mutation detection of all samples by the MASS-PCR and ARMS-PCR methods, and the results were summarized in Table 2. Results showed that there were 63 and 56 positive samples detected by MASS-PCR and ARMS-PCR, respectively. Besides, the kappa value was 0.821, with a significant difference (P<0.001). Although MASS-PCR showed high consistency with ARMS-PCR and Sanger sequencing, 17 samples showed different results, accounting for 5.8% of the total samples. Therefore, the mutation statuses of all samples were further confirmed by Sanger sequencing. Sanger sequencing detected a total of 62 samples that carried the common mutations. The MASS-PCR and Sanger sequencing showed a greater consistency rate (kappa value =0.929, P<0.001), confirming the accuracy of MASS-PCR. Also, we also analyzed the consistency rate between ARMS-PCR and Sanger sequencing. Their consistency rate was 0.830 (Table 3), which was similar to that between MASS-PCR and ARMS-PCR.
Table 2
| Methods | MASS-PCR | Kappa value | P value | ||
|---|---|---|---|---|---|
| Positive | Negative | ||||
| ARMS-PCR | Positive | 51 | 5 | 0.821 | <0.001 |
| Negative | 12 | 225 | |||
| Sanger sequencing | Positive | 59 | 3 | 0.929 | <0.001 |
| Negative | 4 | 227 | |||
MASS-PCR, mutation-selected amplification specific system PCR; ARMS-PCR, amplification refractory mutation system PCR.
Table 3
| Methods | ARMS-PCR | Kappa value | P value | ||
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Sanger sequencing | Positive | 51 | 11 | 0.830 | <0.001 |
| Negative | 5 | 226 | |||
ARMS-PCR, amplification refractory mutation system PCR.
By comparing the mutation statuses among the three detection methods, a total of 20 samples were revealed to be inconsistent. We summarized them in Figure 1. Among these samples, 11 samples were positive by the MASS-PCR and Sanger sequencing while negative by ARMS-PCR. Besides, 3 samples were detected to be positive in MASS-PCR and ARMS-PCR while negative in Sanger sequencing.
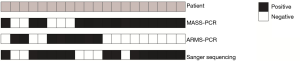
Receiver operating characteristic curve (ROC curve) of MASS-PCR
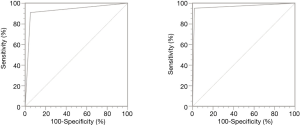
To further assessed the sensitivity and specificity of MASS-PCR, we drew the ROC curve with the reference of ARMS-PCR and Sanger sequencing, respectively. As shown in Figure 2, the areas under curve (AUC) were both larger than 0.9, irrespectively the reference, indicating the high sensitivity and specificity of MASS-PCR.
Discussion
According to histologic features, lung cancer is mainly divided into NSCLC and SCLC, and NSCLC accounts for more than 80% of all lung cancer patients. Recently, with the fast development of genomic medicine, many oncogenic driver mutations have been identified. The common mutations are mostly found in NSCLC while infrequent in SCLC. Among these mutations, EGFR mutation has the highest occurrence rate with 10–15% frequency in NSCLC (2,11), and the frequency was much higher in East Asian patients. In our study, we recruited 293 lung cancer cases with only 6 SCLC cases, and only 1 SCLC case was detected to harbor mutation. Based on the identified oncogenic driver mutations, lung cancer patients would be recommended to receive targeted therapy.
Clinically, before the targeted therapy, the patients need to detect if they harbor the gene mutations that had corresponding targeted drugs. Currently, Sanger sequencing and ARMS-PCR are the main technologies to determine the genotype in the clinic (12). Sanger sequencing (also named as direct sequencing), uses the target DNA single-strand as template to synthesize a new strand upon the catalysis of DNA polymerase. When the fluorescently labeled dideoxynucleotide is selectively synthesized into a new strand, the new strand synthesis is terminated, thus obtaining a series of DNA fragments with varying lengths. Subsequently, electrophoresis was performed to purify the PCR products, then conducting DNA sequencing (13). ARMS-PCR based on the principle that the 3'-terminal nucleotides of the PCR primer must be complementary to its target sequence for efficient amplification. In the amplification system, allele-specific PCR primers and probes labeled with fluorescence are present, and specific fluorescent signals would be generated to indicate the mutant- or wild-type genes (10,14). Compared with Sanger sequencing, ARMS-PCR has a high sensitivity, high specificity and time-saving methods to detect mutations. Although the detection limit of ARMS-PCR is 1%, much lower than that of Sanger sequencing, Sanger sequencing is recognized as the gold standard in the mutation detection, because the positive result detected by Sanger sequencing is a real positive statue. Therefore, Sanger sequencing was used to verify the mutation results in our study. The main limitation of ARMS-PCR is that the determination of mutation status could be subjective when the peak shapes produced by mutant- and wild-type genes are different (15). To solve this defect, MASS-PCR appeared. MASS-PCR is developed based on ARMS-PCR. Only the mutant gene emits a fluorescence peak in the cyclic amplification, and the amplification of wild-type templates is suppressed. Thus, it is considered a positive result as long as the fluorescence peak is generated, reducing the subjectivity on the final determination of detection results (10).
In our study, we compared the performance of MASS-PCR and ARMS-PCR in 293 lung cancer cases. Results revealed a high consistency rate between the two methods. MASS-PCR had a higher consistency rate with Sanger sequencing than with ARMS-PCR. Further analyzing the samples with inconsistent results, we found that 11 of 20 samples were verified to harbor mutations by Sanger sequencing. These samples also detected to be positive by MASS-PCR but not ARMS-PCR. For the rest 9 samples that were detected to be negative by Sanger sequencing, 3 samples were showed positive results in both MASS-PCR and ARMS-PCR. The negative results revealed by Sanger sequencing in these 3 samples might be caused by the false negative. These results demonstrated the higher accuracy and sensitivity of MASS-PCR. The ROC curves also revealed the high sensitivity and specificity of MASS-PCR. Our findings were in line with the work of Zhu et al. (10).
Although we aimed to compare the accuracy and sensitivity of ARMS-PCR and MASS-PCR in both NSCLC and SCLC, only 6 patients were SCLC in our study. Furthermore, the common mutations usually occurred in NSCLC rather than in SCLC. Among the 6 SCLC patients, only 1 SCLC patient was detected to harbor mutation by Sanger sequencing. In the mutation detection of this sample, MASS-PCR assay revealed the same positive result as Sanger sequencing, while, ARMS-PCR assay showed a negative result. This result suggested the higher accuracy and sensitivity of MASS-PCR than ARMS-PCR in the mutation detection of SCLC patients. However, the sample size of SCLC patients was too small, and more SCLC samples must verify our findings further.
In conclusion, we proved that MASS-PCR assay is reliable in the detection of gene mutations with high accuracy, sensitivity, and specificity. Therefore, MASS-PCR could be used in clinical practice to detect gene mutations in lung cancer patients.
Acknowledgments
First, we are supposed to express our gratitude for the support and suggestion in this study from staffs at molecular diagnosis. Second, we thank the technical support from Shanghai Tongshu Biotechnology Co., Ltd.
Funding: This study was funded by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.10.37). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This retrospective study was approved by the board-certified Research Ethics Committee of Sun Yat-sen University Cancer Center (No. B2017-50) and the Institute Research Medical Ethics Committee of Sun Yat-sen University Cancer Center (No. B2018-011 and GZR2019-302). Written informed consent was obtained from all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ettinger DS, Wood DE, Aisner DL, et al. Non-small cell lung cancer, version 5.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2017;15:504-35. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Li T, Kung HJ, Mack PC, et al. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Oncol 2013;31:1039-49. [Crossref] [PubMed]
- Ortiz-Cuaran S, Scheffler M, Plenker D, et al. Heterogeneous mechanisms of primary and acquired resistance to third-generation EGFR inhibitors. Clin Cancer Res 2016;22:4837-47. [Crossref] [PubMed]
- Osmani L, Askin F, Gabrielson E, et al. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol 2018;52:103-9. [Crossref] [PubMed]
- Bakker E. Is the DNA sequence the gold standard in genetic testing? Quality of molecular genetic tests assessed. Clin Chem 2006;52:557-8. [Crossref] [PubMed]
- Liang C, Wu Z, Gan X, et al. Detection of rare mutations in EGFR-ARMS-PCR-negative lung adenocarcinoma by sanger sequencing. Yonsei Med J 2018;59:13-9. [Crossref] [PubMed]
- Zhu Y, Guo Z, Liu Y, et al. A novel ARMS-based assay for the quantification of EGFR mutations in patients with lung adenocarcinoma. Oncol Lett 2018;15:2905-12. [PubMed]
- Ogino S, Kawasaki T, Brahmandam M, et al. Sensitive sequencing method for KRAS mutation detection by pyrosequencing. J Mol Diagn 2005;7:413-21. [Crossref] [PubMed]
- Zhu J, Zhao Y, Liu M, et al. Developing a new qPCR-based system for screening mutation. Small 2019;15:e1805285. [Crossref] [PubMed]
- Mayekar MK, Bivona TG. Current landscape of targeted therapy in lung cancer. Clin Pharmacol Ther 2017;102:757-64. [Crossref] [PubMed]
- Xu S, Duan Y, Lou L, et al. Exploring the impact of EGFR T790M neighboring SNPs on ARMS-based T790M mutation assay. Oncol Lett 2016;12:4238-44. [Crossref] [PubMed]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 1977;74:5463-7. [Crossref] [PubMed]
- Machnicki MM, Glodkowska-Mrowka E, Lewandowski T, et al. ARMS-PCR for detection of BRAF V600E hotspot mutation in comparison with Real-Time PCR-based techniques. Acta Biochim Pol 2013;60:57-64. [Crossref] [PubMed]
- Liu J, Zhao Z, Sun M, et al. The sensitive detection of telomerase reverse transcriptase promoter mutation by amplification refractory mutation system-PCR. Genet Test Mol Biomarkers 2016;20:90-3. [Crossref] [PubMed]


