
LncRNA CASC9 regulates cell proliferation, apoptosis and cell cycle via sponging miR-145-5p in colon cancer cells
Introduction
Colorectal cancer, including colon cancer (CC) and rectal cancer (RC), remains the third highest cancer-related incidence and mortality rate (1). At present, surgical resection and chemotherapy, which are the primary strategies for treating CC, have made substantial progress in recent years (2). However, limited advance has been made in cure rates and long-term survival of CC patients in the past several decades. Therefore, more research should be done to understand the progress of CC and discover effective therapeutic strategies for the treatment of CC.
Long non-coding RNAs (lncRNAs) are important regulatory factors in tumor development and progression (3-5). LncRNAs have complicated functions with multiple mechanisms (6). Mounting evidence indicates the existence of a network of lncRNA-microRNA (miRNA) pathways, and the lncRNA-miRNA-gene interaction patterns have been systematically characterized in cancers. A comprehensive sponge regulatory network mediated by lncRNA has also shown in prostate cancer (7,8) and hepatocellular carcinoma (9). In CRC, lncRNA-PNUTS serves as a competitive sponge for miR-205, suppressing cell growth through destabilizing HuR protein (10). LINC00336 inhibits ferroptosis in lung cancer cells by acting as a competing miR6852 (11). LncRNA Cancer Susceptibility Candidate 9 (CASC9) is known to be an oncogene and is significantly up-regulated in various cancers (12-15). In hepatocellular carcinoma cells, CASC9 interacts with RNA binding protein heterogeneous nuclear ribonucleoprotein L (HNRNPL) and regulates AKT signaling and DNA damage sensing (16,17). In addition, CASC9 is a potential prognosis marker and therapeutic target in esophageal cancer (18). Studies show that the up-regulation of CASC9 promotes tumorigenesis (19) and metastasis (20) of esophageal squamous, doxorubicin-resistant in breast cancer (21). However, the role of CASC9 in CC has been less reported.
One of main the mechanisms through which lncRNAs function in tumor is acting as microRNA sponges. MicroRNAs are short non-coding RNAs that affect gene expression through targeting and regulating mRNAs. MiR-145-5p is a tumor suppressor and closely related to the prognosis of cancer patients (22-25). MiR-145-5p was also identified significantly associated with overall survival and disease-free survival of patients with glioblastoma or gastric cancer (26-28). In CC, miR-145-5p is down-regulated (29), however, population-based data show that miR-145-5p is high-expressed at advanced stage of CC (30). Therefore, we are interested in investigating the role of miR-145-5p in CC.
In this study, we aim to explore the role of CASC9 as a miRNA sponge for miR-145-5p in CC and to provide a new understanding on CASC9 and miR-145-5p in cancers.
Methods
Patients and tissue sample
Thirty samples and normal adjacent tissues were collected from patients [stage I/II (n=17) and III/IV (n=13)] after surgery in the First Affiliated Hospital of Hebei North University, and stored at −80 °C. This study was authorized by the Ethics Committee of the First Affiliated Hospital of Hebei North University (NO. HNU20150703324), and all patients signed an informed consent.
Cell culture
Human intestinal epithelial cells (HIEC, CRL-3266) and human CC cell lines [(LoVo (CCL-229), HCT116 (CCL-247), SW480 (CCL-228), HT29 (HTB-38)] were obtained from the American Type Culture Collection. The HIEC, LoVo, HCT116 and SW480 cells were grown in RPMI-1640 (61870044, Gibco, ThermoFisher, USA) containing 10% FBS (16140071, Invitrogen, ThermoFisher, USA), 1% penicillin/streptomycin (15070063, Gibco, ThermoFisher, USA) at 37 °C with 5% CO2. HT29 cells were grown in McCoy’s 5A (Modified) Medium (16600108, Invitrogen, ThermoFisher, USA) supplemented with 20% FBS (16140071, Invitrogen, ThermoFisher, USA), 1% penicillin/streptomycin (15070063, Gibco, ThermoFisher, USA).
Cell transfection
The siRNA targeted toward human CASC9 (30) (siCASC9: 5'-GCCUGUGAUAGCAGAACAATT-3') and a non-specific negative control siRNA (siNC: 5'-UUCUUCGAACGUGUCACGUTT-3'), miRNAs inhibitor control and miR-145-5p inhibitor were purchased from Gene Pharma Co., Ltd. (Shanghai, China). 100 pmol of siRNA or miRNA inhibitor were respectively transfected into the cells (4×105 cells/well) by using 500 µL Opti-MEM (11058021, Invitrogen, USA) containing 5µL lipofectamine® 2000 (11668019, Invitrogen, USA) at room temperature for 5min. After incubation at 37 °C for 24 h, subsequent functional detections were performed.
RNA isolation and qualitative real time PCR analysis
Tumor specimens and corresponding adjacent normal tissues were disrupted using liquid nitrogen and homogenized using Trizol reagent (15596018, Invitrogen, Thermofisher, USA) at 4 °C. Qiagen RNeasy Plus Mini Kit (74134, Qiagen, Germany) was used to isolate the total RNA from the tissue samples at 4 °C. For isolation of RNA in the cells, the cells were homogenized using TRIzol reagent and isolated by chloroform and islpropanol at 4 °C. The concentration of RNA was measured by using NanoDrop 8000 (ND-8000-GL, Thermo Scientific, USA).
For the quantification of mRNA, cDNA was prepared by using PrimeScript™ II 1st Strand cDNA Synthesis Kit (6210B, Takara, Japan). SYBR® Green PCR Master Mix (4312704, ABI, USA) and Bio-Rad CFX 96 Touch Real-Time PCR Detection System (1855196, Bio-Rad, China) were used for qRT-PCR, with GAPDH served as endogenous control. QRT-PCR parameters were as follows: pre-denaturation at 95 °C for 5 min, then 40 cycles of denaturation (at 95 °C for 30 s), annealing (at 60 °C for 30 s), and extension (at 72 °C for 30 s). For the quantification of microRNA, the cDNA was prepared by One step miRNA RT kit (D1801, HaiGene, China). SYBR Green qPCR kit for miR-145-5p (AP01411, HaiGene, China) and for U6 snRNA (AP02055, HaiGene, China) were then used for qRT-PCR, with U6 snRNA served as an endogenous control. QRT-PCR parameters were as follows: at 95 °C for 5 min, 40 cycles of denaturation (at 95 °C for 15 s), annealing (at 60 °C for 30 s), and extension (at 70 °C for 10 s). 2-∆∆CT was used to calculate the relative expression levels. All primers used in qRT-PCR were shown in Table 1. The primers used in qRT-PCR for miR-145-5p were designed according to one previous study (31).
Table 1
| Primer name | Forward | Reverse |
|---|---|---|
| CASC9 | TTGGTCAGCCACATTCATGGT | AGTGCCAATGACTCTCCAGC |
| MiR-145-5p | TCTTGTCATAAAGGTCCTTAGG | GTGCAGGGTCCGAGGT |
| GAPDH | GGAGCGAGATCCCTCCAAAAT | GGCTGTTGTCATACTTCTCATGG |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
CCK-8 assay
The cells (5×103 cells/well) were cultured in 96-well culture dishes. After culturing at 37 °C for 24, 48 and 72 h, CCK-8 solution (70-CCK801, MultiSciences, China) was separately added into the dishes (10 µL/well) to continue the incubation of the cells at 37 °C for 4 h in the dark. Absorbance values were measured using a microplate reader (1681135, Bio-rad, China) at 450 nm.
Cell colony formation assay
The cells (1,000 cells/well) were seeded into 60 mm plates and cultured for 7 d. Afterwards, colonies were fixed by 4% paraformaldehyde (30525-89-4, Aladdin, China) for 20 min and stained by 0.5% crystal violet (548-62-9, Aladdin, China) at room temperature for 1 h. The picture of colonies was directly took by a camera. The colonies (diameter >0.5 mm) were counted by Image J software v.1.48 (National Institutes of Health, USA). The experiments were conducted in triplicate
Western blotting
The cells were gently washed by 1× PBS twice and treated by 1% NP-40 (FNN0021, Invitrogen, ThermoFisher, USA) supplemented with halt protease inhibitor cocktail (78429, Thermo Scientific, ThermoFisher, USA) on ice and proteins were collected by centrifugation at 1,000 g, 4 °C for 30 min. The concentration of the proteins was measured by Pierce™ BCA Protein Assay Kit (23225, Thermo Scientific, ThermoFisher, USA). Next, 25 µg protein extraction were separated on 10% SDS-PAGE gel and then transferred to PVDF membranes (LC2002, Invitrogen, Thermofisher, USA). The membranes were washed by 1xTBST (50 mM Tris, 150 mM NaCl and 2% Tween-20; pH 7.5) for three times at room temperature and then incubated with primary antibodies (Anti-Ki-67 (ab15580, 1:2,000, Abcam, UK), Anti-PCNA (ab92552, 1:2,000, Abcam, UK) and Anti-GAPDH (ab181602, 1:5,000, Abcam, UK)) at 4 °C overnight. Next, the membranes were further incubated with anti-rabbit IgG antibody (1:2,000, 7074, Cell Signaling Technology, USA) at 4 °C overnight and with SignalFire™ ECL reagent (6883, Cell Signaling Technology, USA) for 1min at room temperature in the dark, and then exposed to X-ray. Images were captured by ImageQuant ECL Imager (28-9605-63, GE Healthcare, USA) and analyzed by using Image J software v.1.48. GAPDH was used for normalization.
Flow cytometry
For evaluation of cell-cycle arrest, the cells (1×106–5×106 cells) were digested by trypsin (25300054, Gibco, Thermofisher, USA) for 2min at 37 °C and collected by centrifugation at 450 g, 4 °C for 5 min. After removing the supernatant, the cells were fixed by 70% ethanol (64-17-5, Aladdin, China) at 4 °C for 2 h and then treated by 1 µg/mL propidium iodide (PI, 25535-16-4, Aladdin, China) in 1 mL PBS at 4 °C for 30 min. Then, FACSCalibur flow cytometer (342973, BD Biosciences, USA) was used for determining cell cycle, and data were analyzed by BD FACSCanto™ system software v2.4 (646602, BD Biosciences, USA).
Annexin-V kit (70-AP101-100-AVF, MultiSciences, China) was used to detect cell apoptosis. After trypsinization, the cells were washed twice by cold 1× PBS. After centrifugation at 450 g, 4 °C for 5 min, the cells (1×106–5×106 cells) were resuspended in 300 µL binding buffer supplemented with 5 µL Annexin-V-FITC solution and incubated for 15 min at room temperature in the dark. The nuclei were stained by 5 µL PI (25535-16-4, Aladdin, China) for 5 min, followed by adding of 200 µL binding buffer. After that, FACSCalibur flow cytometer (342973, BD Biosciences, USA) were used to determine cell apoptosis. BD FACSCanto™ system software v2.4 (646602, BD Biosciences, USA) was used for data analysis.
Dual-luciferase reporter assay
The binding sites for miR-145-5p in lncRNA CASC9 was predicted by the StarBase Software (http://starbase.sysu.edu.cn/). CASC9 wild type sequence and mutation sequence containing miR-145-5p target site were purchased from Tsingke Co., Ltd. and respectively cloned into luciferase reporter gene vector (pmirGLO, E1330, Promega, USA) as CASC9-WT reporter plasmid and CASC9-MUT luciferase reporter plasmid. Briefly, reporter plasmids CASC9-WT and CASC9-MUT were separately transfected into HCT116 cells or SW480 cells. MiRNA-145-5p inhibitor was also transfected into cells by luciferase reporter system. After incubation for 48 h, the luciferase activity was determined by dual-luciferase assay system (E1910, Promega, USA) and Microplate Luminometer (11300010, Berthold, Germany). The firefly luciferase activity was normalized against renilla luciferase activity and presented as mean ± SD.
Statistical analysis
SPSS 16.0 was used for statistical analyses. The data were shown as mean ± standard deviation (SD). Student’s t-test (two-tailed) and one-way ANOVA, followed by Dunnett’ s post hoc test, were performed for statistical analyses. The difference was considered to be significant if P<0.05.
Results
LncRNA CASC9 is up-regulated in CC tissues and cells
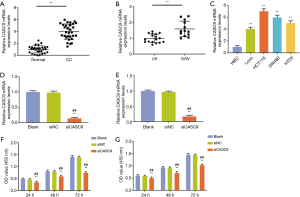
The expression of lncRNA CASC9 in tumor-adjacent CC tissues and normal colonic tissues from CC patients was determined. The results showed that CASC9 was obviously up-regulated in CC tissues (P<0.001), and that the level of CASC9 was higher in CC tissues derived from patients at III/IV stages than those at I/II stages (P<0.001, Figure 1A,B). In addition, the level of lncRNA CASC9 was also evaluated in HIEC cells and LoVo, HCT116, SW480 and HT29 cells. The results demonstrated that CASC9 was up-regulated in CC cell lines than in HIEC cells (P<0.001, Figure 1C), with CASC9 level higher in HCT116 and SW480 cells than that in other CC cell lines, thus, HCT116 and SW480 cells were used for further research.
Knockdown of CASC9 inhibits cell growth, promotes cell apoptosis and arrests cell cycle in CC cells
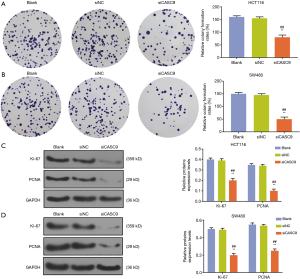
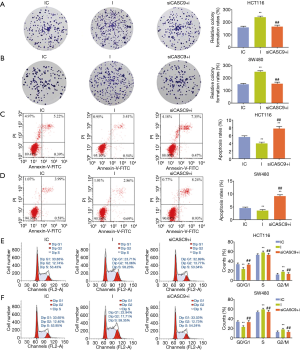
The expression level of CASC9 in HCT116 and SW480 was significantly down-regulated by siCASC9 (both P<0.001, Figure 1D,E). CCK-8 assays showed that the cell viabilities of HCT116 and SW480 cells were decreased by knocking down CASC9 (both P<0.001, Figure 1F and G). Cell colony formation assays in HCT116 and SW480 cells also confirmed that silencing CASC9 suppressed cell growth (both P<0.001, Figure 2A,B). The protein expressions of Ki67 and PCNA protein were determined by western blotting, the result of which showed that the levels of Ki-67 and PCNA in HCT116 and SW480 cells were significantly down-regulated by siCASC9 (both P<0.001, Figure 2C,D).
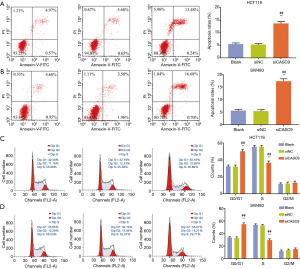
The effects of siCASC9 on cell apoptosis and cycle in HCT116 and SW480 cells were evaluated by flow cytometry, and we found that knockdown of CASC9 caused significant increase of cell apoptosis (both P<0.001, Figure 3A,B). The relative number of cells at G0/G1 phase was increased, while those at S phase was decreased after interference with siCASC9 (both P<0.001, Figure 3C,D).
CASC9 is identified as a molecular sponge for miR-145-5p
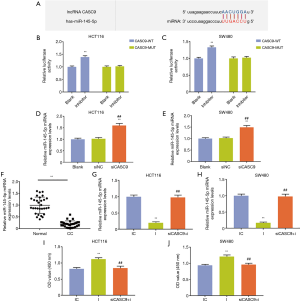
Bioinformatics analysis revealed the binding site for miR-145-5p in CASC9 and indicated a potential link between lncRNA CASC9 and miR-145-5p (Figure 4A). The luciferase activity in HCT116 cells transfected with CASC9-WT was observably increased by miR-145-5p inhibitor (P<0.001), while there was no significant alteration in HCT116 cells transfected with CASC9--MUT reporter (Figure 4B) was observed, moreover, similar results were shown in SW480 cells (Figure 4C). Next, the expression level of miR-145-5p was measured by qRT-PCR, and we observed that knockdown of CASC9 up-regulated the level of miR-145-5p in HCT116 and SW480 cells (both P<0.001, Figure 4D,E). Comparison on the expression level of miR-145-5p in tumor-adjacent normal colonic tissues with that in CC tissues confirmed that miR-145-5p was significantly down-regulated in CC tissues (P<0.001, Figure 4F). Further experiments in HCT116 and SW480 cells showed that the down-regulated level of miR-145-5p by miR-145-5p inhibitor could be rescued by knocking down CASC9 (both P<0.001, Figure 4G,H).
CASC9 regulates cell growth, cell apoptosis and cell cycle arrest via modulating miR-145-5p in CC cells
To further verify the effects of CASC9 on CC cells through miR-145-5p, we evaluated the cell proliferation ability, cell apoptosis and cell cycle arrest in HCT116 and SW480 cells after transfection with miR-145-5p inhibitor or co-transfection with miR-145-5p inhibitor and siCASC9. The results showed that miR-145-5p inhibitor increased the cell viability, while siCASC9 abrogated the effect of miR-145-5p inhibitor on the cells (both P<0.001, Figure 4I,J). Additionally, knockdown of CASC9 also abrogated the increase of cell colony formation rates caused by miR-145-5p inhibitor in HCT116 and SW480 cells (both P<0.001, Figure 5A,B). Flow cytometry results confirmed that the increase of cell apoptosis rates caused by the inhibition of miR-145-5p was also rescued by siCASC9 (both P<0.001, Figure 5C,D). Moreover, miR-145-5p inhibitor increased the cells at S phase but was compromised by siCASC9 in HCT116 and SW480 cells (both P<0.001, Figure 5E,F).
Discussion
LncRNAs are dysregulated in various cancers, and they promote tumor initiation and progression (32). In this study, we found that lncRNA CASC9 acts as an oncogene to regulate cell proliferation, apoptosis and cycle through modulating miR-145-5p in CC cells.
In this study, the level of CASC9 was found higher in CC tissues and cell lines. Moreover, CASC9 was high-expressed in CC tissues at advanced stage, indicating that CASC9 may be related to the prognosis of CC, which is consistent with a previous study, in which higher CASC9 expression is shown to be related to shorter survival in OC patients (33). We also found that knockdown of CASC9 inhibits cell growth, promotes cell apoptosis and cell cycle arrest in CC cells, which is consistent with the previous study that CASC9 is high-expressed in esophageal squamous (34). Generally, increased cell proliferation and inhibition of cell apoptosis cause tumor formation, while inhibition of cell proliferation and induction of cell apoptosis could suppress the development of tumor. Thus, we consider lncRNA CASC9 to be an oncogene in CC.
LncRNA-miRNA-gene interactions contribute to the development of cancers. As a miRNAs sponge, CASC9 affects the development of various cancers through targeting multiple miRNAs, for example, in hemangioma endothelial cells, lncRNA CASC9 regulates cell migration and invasion by targeting miR-125a-3p/Nrg1 (35), while in breast cancer cells, lncRNA CASC9 positively affects the expression of CHK1 through sponging the miR-195/497 cluster (36). Research also showed that in glioma, CASC9, miR-519b and STAT3 form a positive feedback loop, facilitating tumourigenesis (37). In ovarian cancer, CASC9 regulates LIN7A expression via targeting miR-758-3p, contributing to the malignancy of ovarian cancer cells (38). Therefore, we suspected that CASC9 targets different miRNAs in different cancer types, or has multiple miRNA targets, suggesting that the role of CASC9 is complicated in cancer cells. By performing bioinformatics analysis and dual-luciferase reporter assay, we found that miR-145-5p was the functional target for CASC9 in CC cells, which has not been found in other studies, RNA pull-down analysis still needs to be further performed to verify the interaction between CASC9 and miR-145-5p. Previous studies demonstrated that miR-145-5p was the target for multiple lncRNAs. For example, miR-145-5p is sponged by lncRNA TUG1 in laryngocarcinoma (39), in prostate carcinoma, lncRNA BRE-AS1 interacts with miR-145-5p to regulate cancer cell proliferation (40). LncRNA MALAT1 could also up-regulate the level of NEDD9 through targeting miR145-5p (41,42). Apart from lncRNAs, miR-145-5p can also be sponged by circular RNA in bladder cancer (31). Thus, we consider that miR-145-5p can be affected by different lncRNAs. In this study, we confirmed that lncRNA CASC9 targets miR-145-5p in CC cells, and that the level of miR-145-5p in CC tissues was obviously lower than that in tumor-adjacent normal colonic tissues, which was opposite to the expression pattern of CASC9.
Further experiments also confirmed that CASC9 regulates cell growth, apoptosis and cell cycle arrest via modulating miR-145-5p in CC cells. MiR-145-5p is a tumor suppressor in cancer cells, and it inhibits cell proliferation and migration in bladder cancer (43), sensitizes prolactinoma to bromocriptine (44), affects the differentiation of gastric cancer (45), and inhibits gastric cancer invasiveness through suppressing epithelial-mesenchymal transition (46). Moreover, miR-145-5p is also found to be related to different signaling pathways, such as JNK signaling pathway (47), sp1/NF-κB signaling pathway (48), MAPK and PI3K/AKT pathways (49). In our study, we confirm that miR-145-5p is a tumor suppressor in CC.
Conclusions
In conclusion, lncRNA CASC9 is high-expressed in CC and regulates the cell growth, apoptosis and cycle through sponging miR-145-5p. Our research provides a new understanding on the roles of CASC9 and miR-145-5p in CC.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.10.33). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was authorized by the Ethics Committee of the First Affiliated Hospital of Hebei North University (NO. HNU20150703324), and all patients signed an informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Kuipers EJ, Grady WM, Lieberman D, et al. Colorectal cancer. Nat Rev Dis Primers 2015;1:15065. [Crossref] [PubMed]
- Bill M, Papaioannou D, Karunasiri M, et al. Expression and functional relevance of long non-coding RNAs in acute myeloid leukemia stem cells. Leukemia 2019;33:2169-82. [Crossref] [PubMed]
- Hosono Y, Niknafs YS, Prensner JR, et al. Oncogenic Role of THOR, a Conserved Cancer/Testis Long Non-coding RNA. Cell 2017;171:1559-72.e20. [Crossref] [PubMed]
- Huarte M. The emerging role of lncRNAs in cancer. Nat Med 2015;21:1253-61. [Crossref] [PubMed]
- Dykes IM, Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics Proteomics Bioinformatics 2017;15:177-86. [Crossref] [PubMed]
- Wang P, Ning S, Zhang Y, et al. Identification of lncRNA-associated competing triplets reveals global patterns and prognostic markers for cancer. Nucleic Acids Res 2015;43:3478-89. [Crossref] [PubMed]
- Du Z, Sun T, Hacisuleyman E, et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun 2016;7:10982. [Crossref] [PubMed]
- Bai Y, Long J, Liu Z, et al. Comprehensive analysis of a ceRNA network reveals potential prognostic cytoplasmic lncRNAs involved in HCC progression. J Cell Physiol 2019;234:18837-48. [PubMed]
- Lan Y, Xiao X, He Z, et al. Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res 2018;46:5809-21. [Crossref] [PubMed]
- Wang M, Mao C, Ouyang L, et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ 2019;26:2329-43. [Crossref] [PubMed]
- Gao L, Guo YN, Zeng JH, et al. The expression, significance and function of cancer susceptibility candidate 9 in lung squamous cell carcinoma: A bioinformatics and in vitro investigation. Int J Oncol 2019;54:1651-64. [PubMed]
- Yu X, Lin Y, Sui W, et al. Analysis of distinct long noncoding RNA transcriptional fingerprints in pancreatic ductal adenocarcinoma. Cancer Med 2017;6:673-80. [Crossref] [PubMed]
- Su X, Li G, Liu W. The Long Noncoding RNA Cancer Susceptibility Candidate 9 Promotes Nasopharyngeal Carcinogenesis via Stabilizing HIF1α. DNA Cell Biol 2017;36:394-400. [Crossref] [PubMed]
- Ma P, Zhang M, Nie F, et al. Transcriptome analysis of EGFR tyrosine kinase inhibitors resistance associated long noncoding RNA in non-small cell lung cancer. Biomed Pharmacother 2017;87:20-6. [Crossref] [PubMed]
- Klingenberg M, Groß M, Goyal A, et al. The Long Noncoding RNA Cancer Susceptibility 9 and RNA Binding Protein Heterogeneous Nuclear Ribonucleoprotein L Form a Complex and Coregulate Genes Linked to AKT Signaling. Hepatology 2018;68:1817-32. [Crossref] [PubMed]
- Noh JH, Gorospe M. AKTions by Cytoplasmic lncRNA CASC9 Promote Hepatocellular Carcinoma Survival. Hepatology 2018;68:1675-7. [Crossref] [PubMed]
- Pan Z, Mao W, Bao Y, et al. The long noncoding RNA CASC9 regulates migration and invasion in esophageal cancer. Cancer Med 2016;5:2442-7. [Crossref] [PubMed]
- Gao GD, Liu XY, Lin Y, et al. LncRNA CASC9 promotes tumorigenesis by affecting EMT and predicts poor prognosis in esophageal squamous cell cancer. Eur Rev Med Pharmacol Sci 2018;22:422-9. [PubMed]
- Liang Y, Chen X, Wu Y, et al. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ 2018;25:1980-95. [Crossref] [PubMed]
- Jiang B, Li Y, Qu X, et al. Long noncoding RNA cancer susceptibility candidate 9 promotes doxorubicinresistant breast cancer by binding to enhancer of zeste homolog 2. Int J Mol Med 2018;42:2801-10. [PubMed]
- Lin N, Yao Z, Xu M, et al. Long noncoding RNA MALAT1 potentiates growth and inhibits senescence by antagonizing ABI3BP in gallbladder cancer cells. J Exp Clin Cancer Res 2019;38:244. [Crossref] [PubMed]
- Namkung J, Kwon W, Choi Y, et al. Molecular subtypes of pancreatic cancer based on miRNA expression profiles have independent prognostic value. J Gastroenterol Hepatol 2016;31:1160-7. [Crossref] [PubMed]
- Wen J, Luo K, Liu H, et al. MiRNA Expression Analysis of Pretreatment Biopsies Predicts the Pathological Response of Esophageal Squamous Cell Carcinomas to Neoadjuvant Chemoradiotherapy. Ann Surg 2016;263:942-8. [Crossref] [PubMed]
- Wang K, Chen M, Wu W. Analysis of microRNA (miRNA) expression profiles reveals 11 key biomarkers associated with non-small cell lung cancer. World J Surg Oncol 2017;15:175. [Crossref] [PubMed]
- Zhao H, Shen J, Hodges TR, et al. Serum microRNA profiling in patients with glioblastoma: a survival analysis. Mol Cancer 2017;16:59. [Crossref] [PubMed]
- Li CY, Liang GY, Yao WZ, et al. Identification and functional characterization of microRNAs reveal a potential role in gastric cancer progression. Clin Transl Oncol 2017;19:162-72. [Crossref] [PubMed]
- Zhang Y, Wen X, Hu XL, et al. Downregulation of miR-145-5p correlates with poor prognosis in gastric cancer. Eur Rev Med Pharmacol Sci 2016;20:3026-30. [PubMed]
- Pellatt DF, Stevens JR, Wolff RK, et al. Expression Profiles of miRNA Subsets Distinguish Human Colorectal Carcinoma and Normal Colonic Mucosa. Clin Transl Gastroenterol 2016;7:e152. [Crossref] [PubMed]
- Slattery ML, Herrick JS, Mullany LE, et al. An evaluation and replication of miRNAs with disease stage and colorectal cancer-specific mortality. Int J Cancer 2015;137:428-38. [Crossref] [PubMed]
- Wu Z, Huang W, Wang X, et al. Circular RNA CEP128 acts as a sponge of miR-145-5p in promoting the bladder cancer progression via regulating SOX11. Mol Med 2018;24:40. [Crossref] [PubMed]
- Lin C, Yang L. Long Noncoding RNA in Cancer: Wiring Signaling Circuitry. Trends Cell Biol 2018;28:287-301. [Crossref] [PubMed]
- Chen M, Xu X, Ma H. Identification of oncogenic long noncoding RNAs CASC9 and LINC00152 in oral carcinoma through genome-wide comprehensive analysis. Anticancer Drugs 2019;30:356-62. [Crossref] [PubMed]
- Wu Y, Hu L, Liang Y, et al. Up-regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer 2017;16:150. [Crossref] [PubMed]
- Li X, Chen B, Chi D, et al. lncRNA CASC9 regulates cell migration and invasion in hemangioma endothelial cells by targeting miR-125a-3p/Nrg1. Onco Targets Ther 2019;12:423-32. [Crossref] [PubMed]
- Shao G, Wang M, Fan X, et al. lncRNA CASC9 positively regulates CHK1 to promote breast cancer cell proliferation and survival through sponging the miR195/497 cluster. Int J Oncol 2019;54:1665-75. [PubMed]
- Liu H, Li C, Yang J, et al. Long noncoding RNA CASC9/miR-519d/STAT3 positive feedback loop facilitate the glioma tumourigenesis. J Cell Mol Med 2018;22:6338-44. [Crossref] [PubMed]
- Hu X, Li Y, Kong D, et al. Long noncoding RNA CASC9 promotes LIN7A expression via miR-758-3p to facilitate the malignancy of ovarian cancer. J Cell Physiol 2019;234:10800-8. [Crossref] [PubMed]
- Zhuang S, Liu F, Wu P. Upregulation of long noncoding RNA TUG1 contributes to the development of laryngocarcinoma by targeting miR-145-5p/ROCK1 axis. J Cell Biochem 2019;120:13392-402. [Crossref] [PubMed]
- Chen Z, Zhen M, Zhou J. LncRNA BRE-AS1 interacts with miR-145-5p to regulate cancer cell proliferation and apoptosis in prostate carcinoma and has early diagnostic values. Biosci Rep 2019;39:BSR20182097. [Crossref] [PubMed]
- Xue D, Lu H, Xu HY, et al. Long noncoding RNA MALAT1 enhances the docetaxel resistance of prostate cancer cells via miR-145-5p-mediated regulation of AKAP12. J Cell Mol Med 2018;22:3223-37. [Crossref] [PubMed]
- Yu W, Ding J, He M, et al. Estrogen receptor β promotes the vasculogenic mimicry (VM) and cell invasion via altering the lncRNA-MALAT1/miR-145-5p/NEDD9 signals in lung cancer. Oncogene 2019;38:1225-38. [Crossref] [PubMed]
- Zhang H, Jiang M, Liu Q, et al. miR-145-5p inhibits the proliferation and migration of bladder cancer cells by targeting TAGLN2. Oncol Lett 2018;16:6355-60. [PubMed]
- Ma YX, Zhang H, Li XH, et al. MiR-30e-5p inhibits proliferation and metastasis of nasopharyngeal carcinoma cells by target-ing USP22. Eur Rev Med Pharmacol Sci 2018;22:6342-9. [PubMed]
- Zhou T, Chen S, Mao X. miR-145-5p affects the differentiation of gastric cancer by targeting KLF5 directly. J Cell Physiol 2019;234:7634-44. [Crossref] [PubMed]
- Jiang SB, He XJ, Xia YJ, et al. MicroRNA-145-5p inhibits gastric cancer invasiveness through targeting N-cadherin and ZEB2 to suppress epithelial-mesenchymal transition. Onco Targets Ther 2016;9:2305-15. [PubMed]
- Jin X, Chen X, Hu Y, et al. LncRNA-TCONS_00026907 is involved in the progression and prognosis of cervical cancer through inhibiting miR-143-5p. Cancer Med 2017;6:1409-23. [Crossref] [PubMed]
- Mei LL, Wang WJ, Qiu YT, et al. miR-145-5p Suppresses Tumor Cell Migration, Invasion and Epithelial to Mesenchymal Transition by Regulating the Sp1/NF-κB Signaling Pathway in Esophageal Squamous Cell Carcinoma. Int J Mol Sci 2017;18:1833. [Crossref] [PubMed]
- Liu S, Gao G, Yan D, et al. Effects of miR-145-5p through NRAS on the cell proliferation, apoptosis, migration, and invasion in melanoma by inhibiting MAPK and PI3K/AKT pathways. Cancer Med 2017;6:819-33. [Crossref] [PubMed]


