
Predictions for high COL1A1 and COL10A1 expression resulting in a poor prognosis in esophageal squamous cell carcinoma by bioinformatics analyses
Introduction
Esophageal cancer (EC) is the eighth most common malignancy and the sixth leading cause of cancer mortality worldwide (1). In 2013, EC is responsible for more than 440,000 deaths globally (2). Often by the time of diagnosis, local invasion or metastasis has occurred in EC patients. Although the development of chemoradiotherapy or surgery can improve the prognosis, advanced EC patients frequently develop recurrent disease (3). The current median survival time of advanced or metastatic EC patients without esophagectomy is still less than 1 year (4,5). EC mainly includes two pathological types: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). Incidence of ESCC is declining in most countries during recent decades, but ESCC accounts for the vast majority of global EC cases. Therefore, ESCC is still the most common type of EC (6,7). A number of studies focused on the tumorigenesis, progression and therapeutic effect of ESCC have been carried out in recent years (8,9). Although considerable advances have been made in these studies, focusing on a single gene is still not sufficient to reveal the potential mechanisms responsible for the occurrence and development of ESCC. Investigating the molecular mechanisms and molecular biomarkers for early diagnosis, surveillance and targeted therapy of ESCC by bioinformatics methods are warranted.
High throughput sequencing has been widely used in human cancers. Clinical and sequencing data can be downloaded from various public databases. In the present study, two datasets of ESCC (GSE20347 and GSE70409) from gene expression omnibus (GEO) were analyzed using GEO2R online tool to identify the differentially expressed genes (DEGs). Subsequently, functions and pathways enrichment analyses of DEGs were performed with database for annotation, visualization and integrated discovery (DAVID), and then their protein-protein interaction (PPI) network and cluster analysis were constructed by search tool for the retrieval of interacting genes (STRING). Following prediction of the key DEGs in ESCC, gene expression profiling interactive analysis (GEPIA) was utilized to demonstrate the prediction and analyze the prognosis of ESCC patients.
Methods
Ethics, consent and permissions
All experiments utilizing animal and human samples were approved by the Ethical Committee of Medical Research, The First Affiliated Hospital of Anhui Medical University (No. S2019008). All participants gave informed consent before taking part.
Datasets information
GEO is a database repository of high throughput gene expression data, hybridization arrays, chips and microarrays. Numerous experiments and curated gene expression profiles are available for global users to query and download. In the present study, gene expression profiles GSE20347 and GSE70409 were downloaded from the GEO database. GSE20347 contains 17 tumor tissues and 17 matched normal adjacent tissues from 17 ESCC patients from a high-risk region of China. In the GSE70409 dataset, the tumor and adjacent normal tissues were obtained from 17 male ESCC patients who underwent total esophagectomy without previous cancer treatment.
Identification of differential expression genes
We used GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/), an online tool for analyzing DEGs, to compare tumor and normal samples from the two datasets in order to identify DEGs. Results were reported as a table of genes ordered by significance. In the GEO2R online analysis tool, the data from both datasets were normalized by adjusting the option parameters. We set “apply adjustment to the P values” to “Benjamini & Hochberg (false discovery rate)” and “apply log transformation to the data” to “auto-detect”. Based on the analysis results, genes that met the cut-off criteria of the adjusted P value <0.05 and |log fold change| >2 were defined as DEGs. Venn Diagrams website (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to graphical analyze the distribution of all the DEGs in GSE20347 and GSE70409.
Gene ontology (GO) and pathway enrichment analysis
GO analysis is widely used to annotate genes and gene products, reduce complexity and highlight biological processes (BPs) in genome-wide expression studies. Kyoto encyclopedia of genes and genomes (KEGG) (10) is a database that integrates genomic, chemical and systemic functional information. KEGG pathway analysis is extensively used in molecular interaction, reaction and relation networks. DAVID is an online tool (https://david.ncifcrf.gov/) for functional interpretation of target genes. Both GO and KEGG pathway analysis are integrated in DAVID. BPs, molecular functions (MFs), cellular components (CCs) and pathways of DEGs were analyzed by DAVID, P<0.05 was considered statistically significant.
PPI network and cluster analysis
PPI was evaluated by the STRING, which is an online tool (https://string-db.org) for functional protein association networks. The potential relationship among those DEGs was mapped by STRING. Minimum required interaction score was defined as greater than 0.4. Cluster analysis was performed subsequently to determine the probable key molecules. PPI network was clustered to 3 clusters through k-means clustering.
Expression and prognosis analysis of key DEGs in TCGA database
GEPIA is a web-based tool (http://gepia.cancer-pku.cn/) for cancer and normal gene expression profiling and interactive analyses based on TCGA and GTEx data (11). Boxplot analyses via GEPIA were used to show the expression levels of key DEGs in ESCC tumor tissues and normal tissues. Through GEPIA, Kaplan-Meier curves for disease-free survival were used to evaluate the prognosis of ESCC patients with different DEGs expression levels.
Results
Identification of commonly changed DEGs from different datasets
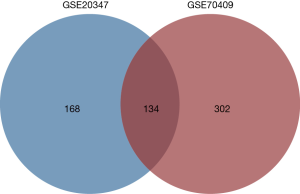
In the present study, a total of 34 ESCC samples and 34 normal samples from two datasets (GSE20347 and GSE70409) were collected for DEGs analysis. Genes that met the cut-off criteria of the adjusted P value <0.05 and |log fold change| >2 were defined as DEGs. GSE20347 contains 17 tumor tissues and 17 matched normal adjacent tissues. 302 DEGs were identified by the GEO2R online analysis tool, including 100 up-regulated DEGs and 202 down-regulated DEGs. In the GSE70409 dataset, the tumor and normal samples were obtained from 17 male ESCC patients. A total of 436 DEGs were found by the GEO2R tool, of which 134 DEGs were up-regulated and 302 DEGs were down-regulated. After integrated bioinformatical analysis on the Venn Diagrams website, 134 commonly changed DEGs were identified from the two datasets, including 33 up-regulated DEGs and 101 down-regulated DEGs (Figure 1).
GO and KEGG pathway enrichment analysis of commonly changed DEGs
For further understanding of 134 commonly changed DEGs, the online tool DAVID was used to analyze the functions and pathways enrichment of these genes (Tables 1,2). GO analysis results were classified into BP group, CC group and MF group. In the BP group, up-regulated commonly changed DEGs were mainly enriched in collagen catabolic process, extracellular matrix (ECM) organization and disassembly, and down-regulated commonly changed DEGs were mainly enriched in peptide cross-linking, epithelial cell and keratinization differentiation. For CC, up-regulated genes were mainly enriched in extracellular region, and down-regulated genes were mainly enriched in extracellular exosome and cornified envelope. GO MF analysis indicated that up-regulated genes were significantly enriched in ECM structural constituent, and down-regulated genes were mostly enriched in serine-type peptidase activity, iron ion binding and structural molecule activity.
Table 1
| Category & term | Annotation | Count | P value | FDR |
|---|---|---|---|---|
| BP | ||||
| GO:0030574 | Collagen catabolic process | 7 | 1.65×10-9 | 2.26 |
| GO:0030198 | ECM organization | 9 | 1.87×10-9 | 2.57×10-6 |
| GO:0001501 | Skeletal system development | 6 | 4.81×10-6 | 6.60×10-3 |
| GO:0022617 | ECM disassembly | 5 | 1.11×10-5 | 1.52×10-2 |
| GO:0001649 | Osteoblast differentiation | 4 | 9.14×10-4 | 1.24 |
| CC | ||||
| GO:0005576 | Extracellular region | 17 | 1.07×10–9 | 1.14×10–6 |
| GO:0031012 | ECM | 9 | 2.51×10–8 | 2.69×10–5 |
| GO:0005615 | Extracellular space | 14 | 1.08×10–7 | 1.16×10–4 |
| GO:0005578 | Proteinaceous ECM | 8 | 2.67×10–7 | 2.86×10–4 |
| GO:0005581 | Collagen trimer | 6 | 4.50×10–7 | 4.82×10–4 |
| MF | ||||
| GO:0005201 | ECM structural constituent | 4 | 2.73×10–4 | 0.30 |
| GO:0005509 | Calcium ion binding | 6 | 1.06×10–2 | 11.07 |
| GO:0004252 | Serine-type endopeptidase activity | 4 | 1.22×10–2 | 12.67 |
| KEGG pathway | ||||
| hsa04512 | ECM-receptor interaction | 5 | 7.87×10–5 | 7.19×10–2 |
| hsa04974 | Protein digestion and absorption | 4 | 1.67×10–3 | 1.52 |
| hsa04510 | Focal adhesion | 5 | 2.09×10–3 | 1.90 |
GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes; DEGs, differentially expressed genes; ESCC, esophageal squamous cell carcinoma; FDR, false discovery rate; BP, biological process; ECM, extracellular matrix; CC, cellular component; MF, molecular function.
Table 2
| Category & term | Annotation | Count | P value | FDR |
|---|---|---|---|---|
| BP | ||||
| GO:0030216 | Keratinocyte differentiation | 7 | 3.92×10–6 | 5.74×10–3 |
| GO:0031424 | Keratinization | 5 | 1.42×10–4 | 0.21 |
| GO:0018149 | Peptide cross-linking | 5 | 1.67×10–4 | 0.24 |
| GO:0030855 | Epithelial cell differentiation | 5 | 6.11×10–4 | 0.89 |
| GO:0008544 | Epidermis development | 5 | 1.27×10–3 | 1.84 |
| CC | ||||
| GO:0070062 | Extracellular exosome | 46 | 5.05×10–13 | 5.70×10–10 |
| GO:0001533 | Cornified envelope | 6 | 4.44×10–6 | 5.02×10–3 |
| GO:0016324 | Apical plasma membrane | 9 | 1.62×10–4 | 0.18 |
| GO:0005615 | Extracellular space | 18 | 6.99×10–4 | 0.79 |
| MF | ||||
| GO:0008236 | Serine-type peptidase activity | 5 | 4.01×10–4 | 0.52 |
| GO:0005506 | Iron ion binding | 6 | 1.57×10–3 | 2.02 |
| GO:0005198 | Structural molecule activity | 7 | 2.44×10–3 | 3.12 |
| KEGG pathway | ||||
| hsa00982 | Drug metabolism—CYP450 | 4 | 4.39×10–3 | 4.43 |
| hsa00590 | Arachidonic acid metabolism | 3 | 3.70×10–2 | 32.21 |
GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes; DEGs, differentially expressed genes; ESCC, esophageal squamous cell carcinoma; FDR, false discovery rate; BP, biological process; CC, cellular component; MF, molecular function; CYP450, cytochrome P450.
Based on the results of KEGG pathway enrichment analysis, the up-regulated commonly changed DEGs were particularly enriched in ECM-receptor interaction, focal adhesion, protein digestion and absorption, and the down-regulated commonly changed DEGs were mostly enriched in drug and arachidonic acid metabolism.
Key DEGs identification by PPI network and cluster analysis
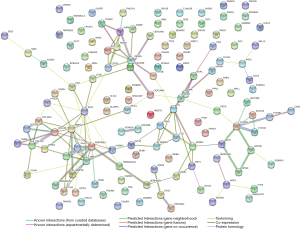
After investigating the potential functions and pathways of 134 commonly changed DEGs, we analyzed the PPI network of these DEGs (Figure 2). The minimum required interaction score was defined as greater than 0.4 (12). To determine the potential key molecules associated with ESCC, cluster analysis was performed subsequently. PPI network of commonly changed DEGs was clustered to 3 clusters through k-means clustering, and the most significant two clusters (cluster 1 & 2) were shown in Figure 3.
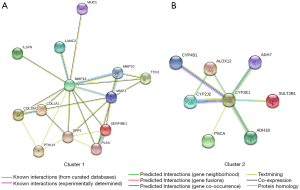
A total of 13 DEGs in cluster 1 and 8 DEGs in cluster 2 were considered as key DEGs. Cluster 1 contained 10 up-regulated key DEGs (COL10A1, COL1A1, LAMC2, MMP1, MMP10, MMP13, PLAU, PTHLH, SERPINE1 and SPP1) and 3 down-regulated DEGs (IL1RN, MUC1 and TCN1). All 8 key DEGs in cluster 2 were down-regulated genes (ADH1B, ADH7, ALOX12, CYP2E1, CYP2J2, CYP4B1, PSCA and SULT2B1). GO and KEGG pathway enrichment analyses of these key DEGs were performed to further evaluate their functions and mechanisms in ESCC (Tables 3,4). GO BP analysis indicated that key DEGs in cluster 1 were mainly enriched in regulation of receptor activity and collagen catabolic process, and key DEGs in cluster 2 were mainly enriched in lipid metabolic process. For CC, key DEGs in both clusters were associated with ECM. For MF, key DEGs in cluster 1 were mainly enriched in metalloendopeptidase activity, and key DEGs in cluster 2 were mainly enriched in ion binding. KEGG pathway enrichment analysis showed that cluster 1 was mainly associated with complement and coagulation cascades, and cluster 2 was mainly related to metabolism. KEGG pathway enrichment analysis.
Table 3
| Category & term | Annotation | Count | P value | FDR |
|---|---|---|---|---|
| BP | ||||
| GO:0010469 | Regulation of receptor activity | 2 | 2.40×10–3 | 2.26 |
| GO:0030574 | Collagen catabolic process | 2 | 1.04×10–2 | 9.42 |
| CC | ||||
| GO:0005578 | Proteinaceous ECM | 4 | 1.69×10–4 | 0.11 |
| GO:0005615 | Extracellular space | 5 | 1.65×10–3 | 1.11 |
| GO:0031012 | ECM | 3 | 1.93×10–3 | 1.30 |
| MF | ||||
| GO:0004222 | Metalloendopeptidase activity | 3 | 1.58×10–3 | 0.86 |
| GO:0005509 | Calcium ion binding | 3 | 2.60×10–2 | 13.45 |
| KEGG pathway | ||||
| ocu04610 | Complement and coagulation cascades | 2 | 4.85×10–2 | 31.49 |
GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes; FDR, false discovery rate; BP, biological process; CC, cellular component; ECM, extracellular matrix; MF, molecular function.
Table 4
| Category & term | Annotation | Count | P value | FDR |
|---|---|---|---|---|
| BP | ||||
| GO:0006629 | Lipid metabolic process | 4 | 2.72×10–4 | 0.33 |
| GO:0006631 | Fatty acid metabolic process | 3 | 9.28×10–4 | 1.13 |
| GO:0032787 | Monocarboxylic acid metabolic process | 3 | 2.19×10–3 | 2.66 |
| CC | ||||
| GO:0005576 | Extracellular region | 4 | 8.12×10–2 | 42.00 |
| MF | ||||
| GO:0005506 | Iron ion binding | 4 | 1.80×10–5 | 0.019 |
| GO:0046914 | Transition metal ion binding | 5 | 3.15×10–4 | 0.34 |
| GO:0004497 | Monooxygenase activity | 3 | 4.95×10–4 | 0.53 |
| KEGG pathway | ||||
| bta00590 | Arachidonic acid metabolism | 3 | 5.42×10–4 | 0.38 |
| bta01100 | Metabolic pathways | 4 | 1.56×10–2 | 10.33 |
GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes; FDR, false discovery rate; BP, biological process; CC, cellular component; MF, molecular function.
Validation in the TCGA and GTEx projects
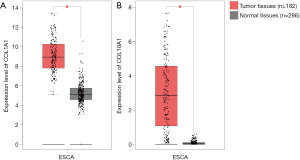
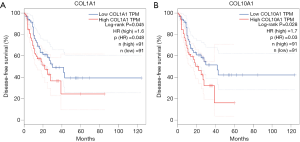
GEPIA is an online tool based on TCGA and GTEx data. To further verify the reliability of the key DEGs in ESCC identified from the two datasets, we queried the expression levels and prognostic information about these key DEGs in ESCC with GEPIA. Among all the 21 key DEGs, 14 DEGs (LAMC2, MMP10, MMP1, SPP1, PLAU, SERPINE1, CYP4B1, ALOX12, ADH7, SULT2B1, ADH1B, CYP2E1, PSCA and IL1RN) were reconfirmed by GEPIA, and the expression trends remained unchanged. In addition, Kaplan-Meier survival analyses for disease-free survival were performed to evaluate the prognosis of ESCC patients with different expression levels of 14 key DEGs. We found that COL1A1 and COL10A1 were significantly increased in ESCC tumor tissues compared with matched normal tissues (Figure 4). Furthermore, disease-free survival for ESCC patients with low COL1A1 or COL10A1 expression was significantly higher than those with high COL1A1 or COL10A1 expression (Figure 5). These results suggested that COL1A1 and COL10A1 might play an important role in the progression of ESCC.
Discussion
Despite considerable studies have been conducted to reveal the etiology and pathogenesis of ESCC in the last few years, the incidence and mortality of EESC is still very high, and the precise mechanism remains unclear. Focusing on a single genetic event or a single cohort study is more likely responsible for this situation. In the present study, we integrated two ESCC datasets from diverse populations, and 134 commonly changed DEGs (33 up-regulated and 101 down-regulated) were identified with multiple bioinformatics methods for the in-depth analysis of these datasets. Subsequently, 134 commonly changed DEGs were enriched by GO and KEGG pathway analysis, and the most two significant clusters with 21 key DEGs were identified using PPI and clustering methods. Following these analyses, 14 key DEGs were reconfirmed by GEPIA which is based on TCGA and GTEx data, and the consistencies in expression trends were maintained. Different DEGs validated in different databases are likely the result of the genomic instability of cancers. Not all mutated genes are referred to driver genes associated with carcinogenesis. Another possibility is that intratumoral heterogeneity might contribute to the significant differences in the molecular mechanisms.
BP analysis of these DEGs was mainly enriched in regulation of receptor activity, collagen catabolic process and lipid metabolism. CCs were mainly related to ECM, and MFs were mostly associated with metalloendopeptidase activity and ion binding. KEGG pathway enrichment analysis showed that these key DEGs were significantly enriched in metabolic pathways, complement and coagulation cascades. Finally, two genes (COL1A1 and COL10A1) in the 14 key DEGs were significantly correlated with disease-free survival in ESCC patients. However, there was no difference in overall survival (data not shown) in patients with different expression levels of COLA1 or COL10A1. Endpoint events of overall survival could be influenced by censoring and various confounders. Disease-free survival provides an advantage to the evaluation of cancer recurrence and progression.
Collagen, a protein that strengthens and supports many tissues in the body, plays an important role in maintaining ECM homeostasis. Most studies so far have shown that ECM are strongly associated with tumor invasion and metastasis (13,14). The presence of collagen in ECM has been known to influence cancer cells. Collagens can induce EMT (epithelial-mesenchymal transition) and related disease progression in many cancers (15,16). Alpha-1 type I collagen is a protein that in humans is encoded by the COL1A1 gene, and alpha-1 type X collagen is another member of the collagen family encoded by the COL10A1 gene. Therefore, we could hypothesize that COL1A1 and COL10A1 might contribute to the invasion and metastasis of ESCC. To test the hypothesis, Kaplan-Meier survival analysis for disease-free survival was performed. We found that high expression of COL1A1 or COL10A1 in ESCC patients was associated with a significant increase in the risk of disease recurrence or progression.
Type I collagen is mainly encoded by the COL1A1 gene, and the most ECM proteins are comprised of fibrillar collagens (17). A recent study has suggested that COL1A1 was up-regulated in colorectal cancer tissues and matched lymph node tissues. COL1A1 overexpression might promote colorectal cancer cell migration through the WNT/PCP pathway (18). Compared with normal tissues, the expression of COL1A1 was also significantly increased in cervical cancer tissues. COL1A1 activation was correlated with low radiosensitivity and anti-apoptosis induced by radiation in cervical cancer cells (19). Conversely, the proliferation and migration of human oral squamous cell carcinoma cells could be inhibited by down-regulating the expression of COL1A1 (20). Similar results have also been reported in gastric cancer (21). These studies suggested that overexpression of COL1A1 could be considered as an important molecular event in a variety of human cancers. Thus, COL1A1 expression may represent an independent biomarker for prediction of prognosis in patients with ESCC.
Type X collagen is a homotrimer which is encoded by the COL10A1 gene. Screening for a panel of RNAs prepared from different cancers and cancer cell lines demonstrated that COL10A1 up-regulated frequently in a variety of cancers. Whereas COL10A1 was restricted expressed or even undetectable in most of normal tissues. COL10A1 can be specifically expressed in the vasculature and tumor microenvironment in breast cancer tissues through immunofluorescence staining with specific antibodies (22). These conclusions were supported by another study. High COL10A1 expression was associated with poor prognosis in patients with ER+/HER2+ breast cancer (23). Besides many different advanced cancers, aberrant expression of COL10A1 can also be detected in precancerous lesions and early tumors. Serum protein concentrations of COL10A1 were markedly increased in adenomas and colorectal cancer cases compared with the control samples. COL10A1 protein levels in serum might be considered as an early potential diagnostic biomarker candidate to identify colorectal cancer and adenoma (24). In our study, as one of the most significant key DEGs, the specificity and sensitivity of COL10A1 in the early diagnosis of ESCC remain to be further investigated.
In summary, we identified several key genes and outcome in ESCC by bioinformatics analyses. A total of 14 key DEGs were selected and validated in different databases. Among these key DEGs, COL1A1 and COL10A1 are significantly associated with the prognosis in ESCC. Therefore, COL1A1 and COL10A1 might be considered as novel potential diagnostic and prognostic biomarkers in patients with ESCC. Further verification experiments are required to confirm the expressions, functions and pathways of these key DEGs.
Acknowledgments
We thank Dr. Cao and Dr. Zhang for offering their help.
Funding: This work was supported by grants from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.11.11). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All experiments utilizing animal and human samples were approved by the Ethical Committee of Medical Research, The First Affiliated Hospital of Anhui Medical University (No. S2019008). All participants gave informed consent before taking part.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- McGuire S. World cancer report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr 2016;7:418-9. [Crossref] [PubMed]
- GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015;385:117-71. [Crossref] [PubMed]
- Lordick F, Mariette C, Haustermans K, et al. Oesophageal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2016;27:v50-7. [Crossref] [PubMed]
- Waddell T, Chau I, Cunningham D, et al. Epirubicin, oxaliplatin, and capecitabine with or without panitumumab for patients with previously untreated advanced oesophagogastric cancer (REAL3): a randomised, open-label phase 3 trial. Lancet Oncol 2013;14:481-9. [Crossref] [PubMed]
- Shah MA, Bang YJ, Lordick F, et al. Effect of fluorouracil, leucovorin, and oxaliplatin with or without onartuzumab in HER2-negative, MET-positive gastroesophageal adenocarcinoma: the METGastric randomized clinical trial. JAMA Oncol 2017;3:620-7. [Crossref] [PubMed]
- Lin Y, Totsuka Y, He Y, et al. Epidemiology of esophageal cancer in Japan and China. J Epidemiol 2013;23:233-42. [Crossref] [PubMed]
- Arnold M, Soerjomataram I, Ferlay J, et al. Global incidence of oesophageal cancer by histological subtype in 2012. Gut 2015;64:381-7. [Crossref] [PubMed]
- Sato Y, Motoyama S, Wakita A, et al. TLR3 expression status predicts prognosis in patients with advanced thoracic esophageal squamous cell carcinoma after esophagectomy. Am J Surg 2018;216:319-25. [Crossref] [PubMed]
- Yang Q, Lin W, Liu Z, et al. RAP80 is an independent prognosis biomarker for the outcome of patients with esophageal squamous cell carcinoma. Cell Death Dis 2018;9:146. [Crossref] [PubMed]
- Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353-61. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102. [Crossref] [PubMed]
- Dahiya S, Saini V, Kumar P, et al. Protein-protein interaction network analyses of human WNT proteins involved in neural development. Bioinformation 2019;15:307-14. [Crossref] [PubMed]
- Lee SW, Kwak HS, Kang MH, et al. Fibroblast-associated tumour microenvironment induces vascular structure-networked tumouroid. Sci Rep 2018;8:2365. [Crossref] [PubMed]
- Bekaert S, Fillet M, Detry B, et al. Inflammation-generated extracellular matrix fragments drive lung metastasis. Cancer Growth Metastasis 2017;10:1179064417745539. [Crossref] [PubMed]
- Grither WR, Divine LM, Meller EH, et al. TWIST1 induces expression of discoidin domain receptor 2 to promote ovarian cancer metastasis. Oncogene 2018;37:1714-29. [Crossref] [PubMed]
- Liu CC, Lin JH, Hsu TW, et al. Collagen XVII/laminin-5 activates epithelial-to-mesenchymal transition and is associated with poor prognosis in lung cancer. Oncotarget 2016;9:1656-72. [PubMed]
- Yamauchi M, Taga Y, Hattori S, et al. Analysis of collagen and elastin cross-links. Methods Cell Biol 2018;143:115-32. [Crossref] [PubMed]
- Zhang Z, Wang Y, Zhang J, et al. COL1A1 promotes metastasis in colorectal cancer by regulating the WNT/PCP pathway. Mol Med Rep 2018;17:5037-42. [PubMed]
- Liu S, Liao G, Li G. Regulatory effects of COL1A1 on apoptosis induced by radiation in cervical cancer cells. Cancer Cell Int 2017;17:73. [Crossref] [PubMed]
- He B, Lin X, Tian F, et al. MiR-133a-3p inhibits oral squamous cell carcinoma (OSCC) proliferation and invasion by suppressing COL1A1. J Cell Biochem 2018;119:338-46. [Crossref] [PubMed]
- Wang Q, Yu J. MiR-129-5p suppresses gastric cancer cell invasion and proliferation by inhibiting COL1A1. Biochem Cell Biol 2018;96:19-25. [Crossref] [PubMed]
- Chapman KB, Prendes MJ, Sternberg H, et al. COL10A1 expression is elevated in diverse solid tumor types and is associated with tumor vasculature. Future Oncol 2012;8:1031-40. [Crossref] [PubMed]
- Brodsky AS, Xiong J, Yang D, et al. Identification of stromal ColXα1 and tumor-infiltrating lymphocytes as putative predictive markers of neoadjuvant therapy in estrogen receptor-positive/HER2-positive breast cancer. BMC Cancer 2016;16:274. [Crossref] [PubMed]
- Solé X, Crous-Bou M, Cordero D, et al. Discovery and validation of new potential biomarkers for early detection of colon cancer. PLoS One 2014;9:e106748. [Crossref] [PubMed]


