
Research progress of cancer stem cells and IL-6/STAT3 signaling pathway in esophageal adenocarcinoma
Introduction
Previous studies showed that only 0.1–0.3% of Barrett’s esophagus (BE) [the critical precancerous stage of esophageal adenocarcinoma (EAC)] developed into EAC (1-3), and about 80–90% of patients with EAC did not have a past medical history of BE (4), but the risk of BE population with EAC was indeed significantly higher, 30–50 times higher than the general population (5). In September 2018, the updated epidemiological data of the National Institutes of Health in the USA showed that about 17,290 cases of EAC were diagnosed between 1975 and 2015 with the death number as high as 15,850 cases, and the 5-year survival rate less than 20%. For locally progressive patients, the main treatment regimen is neoadjuvant chemoradiotherapy combined with surgery (6). In addition, 40% of the patients were diagnosed as too advanced to be removed surgically, resulting in a 5-year survival rate of less than 5% in this case (7). At present, it is believed that the causes of poor prognosis, low survival rate, recurrence, metastasis, chemotherapeutic resistance and radiotherapeutic resistance of malignant tumors may be closely related to the existence of cancer stem cells (CSCs).
Content
The origin of BE, the critical stage of EAC
Understanding the relationship between treatment and relapse is important to improve treatment outcomes and reduce failure of all kinds of cancers. EAC is a suitable disease model to explore the activities and mechanisms of CSCs in cancer, because one of the critical developmental stages has involved characteristic cellular morphological changes, which may suggest the involvement of CSCs. In the disease model, EAC derived from normal esophagus passes through several developmental stages. Chronic exposure of normal esophagus to gastric reflux leads to BE, BE to low grade dysplasia (LGD), LGD to high-grade dysplasia (HGD) and finally HGD to EAC. BE has been observed replacing their squamous epithelial cells with columnar epithelial cells in the distal esophagus (8), which highly suggests the involvement of stem cell related activities (9).
No consensus has been reached about the origin of BE in the academic world. The debate of formation of BE could be classified into two hypotheses: trans-commitment and trans-differentiation. At present, the latest view claims that stem cells play important roles in the formation and progression of BE, which may be differentiated from stem cells (10) (Figure 1).
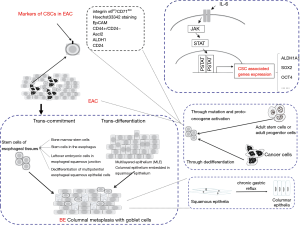
The hypothesis of trans-commitment thinks that BE is directly derived from the stem cell group in the body. Stem cells of esophageal tissues are thought to mainly come from the following four aspects. First, it is from bone marrow stem cells: mesenchymal stem cells promote the differentiation of hematopoietic stem cells (to become matured red blood cells), production of connective tissue, lipid, cartilage and bone, and in specific inducing condition, hematopoietic stem cells can also differentiate into nervous tissue, myocardial cells, skeletal muscle cells and liver cells (11). However, some researchers transplanted bone marrow stem cells to the damaged esophagus where the bone marrow stem cells were observed to differentiate into columnar cells. Second, it is from stem cells in the esophagus: those stem cells are mainly distributed in esophageal squamous epithelial papilla substratum, the neck of the submucosal glandular canal of the esophagus or cardiac gland epithelium (12). Stimulation from the outside environment and growth factor produced by mesenchymal cells in microenvironment could induce the abnormal differentiation of multipotential stem cells which then transform into BE. BE could be induced in the rat model without submucosal glands of the esophagus. It indicated that there might be different location of stem cells inducing the formation of stem cells. Third, it is from the leftover embryonic cells in esophageal squamous junction: leftover embryonic cells in gastro-esophageal junction may undergo opportunistic expansion by penetrating basilar membrane to the apoptotic esophageal squamous epithelium forming BE. Fourth, it is from the dedifferentiation of multipotential esophageal squamous epithelial cells: cell reprogramming means that matured differentiated thesocytes are dedifferentiated to restore totipotency, pluripotent state or the process to become multipotential stem cells. In short, it is the dedifferentiation of mature cells. Cell reprogramming could take part in the occurrence and progression of diseases. Induced pluripotent stem cells (IPSCs) is the typical example of cell reprogramming that mature cells dedifferentiate into embryo stem cell liked multipotential stem cells through cell reprogramming. Cell reprogramming involves the activation of several signaling pathways including BMP/SMAD, WNT/β-catenin, LIF/STAT3, etc. Some researches indicated that squamous cells would be induced to transform into the phenotype of columnar epithelial cells who expressed OCT4 (13), KLF4 (14,15) and other markers of intestinal stem cells through the direct stimulation of bile acid in normal esophageal squamous epithelial cells. In this case, under the stimulation of long-term unhealthy environmental factors like gastric esophageal reflux content, esophageal squamous epithelial cells would dedifferentiate into multi-potential cells who obtained the ability of dedifferentiation through cell reprogramming. After that, the expression of CDX2, MUC2 and other markers of intestinal epithelium would be further targeted and activated so that expression of the marker of squamous epithelium will be down regulated and not detected. At the end, the multi-potential esophageal epithelial cells will differentiate as BE.
The hypothesis of trans-differentiation comes from multilayered epithelium (MLE). MLE, columnar epithelium arranged on the second to the eighth level of squamous epithelium, appears on the BE squamous columnar junction whose surface was covered by visible microvilli under scanning electronic microscopy; meanwhile, MLE expressed CK4 as the characteristics of squamous epithelium and CK19 as the characteristics of columnar epithelium. MLE is the intermediate state from squamous to columnar epithelium during the forming process of BE cellular characteristics. In the normal developmental process of mouse embryo, researchers found that the esophageal overlying squamous epithelium was directly derived from simple columnar epithelium in origin which indicated the existence of reciprocal transformation between squamous and columnar epithelium (16).
Markers of CSCs in EAC
CSCs are a subgroup of cell in solid tumors, with highly efficient proliferation and multidirectional differentiation potential, which is considered the origin of tumorigenesis. The existence of CSCs has confirmed in large number of malignant tumors, including breast cancer, lung cancer, pancreatic cancer, gastric cancer, colorectal cancer, liver cancer, melanoma, and of course, EAC (Figure 1). The identification and isolation of CSCs in EAC have important potential significance for improving the therapeutic efficacy for EAC patients, and targeted CSC therapy may contribute to the ultimate elimination of tumors.
There are relatively few studies on CSCs in EAC. Currently, molecular markers used to identify CSCs in EAC include ALDH1, CD44+/CD24−, ALDH, CD24, integrin α6bri/CD71dim, Ascl2, EpCAM and Hoechst33342 staining. The CSCs of EAC are usually insensitive to 5-fluorouracil and cisplatin in conventional chemotherapy. CD44+/CD24− CSCs isolated from EAC are highly proliferative and spheroid-forming, and are more radiation-resistant than ordinary stem cells. In addition, patients with CD44+/CD24− EAC are less effective in neoadjuvant chemoradiotherapy (17). The EAC cell line resistant to 5-fluorouracil has high expression of CD24 stem cell markers, and CD24 expressing cells have the ability to form into pellets in vitro (18). Studies have shown that OE19 EAC cell lines highly express EpCAM after being resistant to the combined chemotherapy drugs azithromycin, cisplatin and 5-fluorouracil, and form into spheres in serum-free medium and tumors in transplanted nude mice, indicating that the CSC subpopulation of EAC leads to tumor progression and chemotherapy resistance (19). The cells isolated by Hoechst33342 staining are called side population (SP). Drug transporter ABCG2 was the determinant factor of the leakage of Hoechst33342 dye from SP which can lead to the cell leakage of chemotherapeutic drugs, reduce the intracellular concentration of chemotherapy drugs, achieving the effect of chemotherapeutic resistance. The biological behavior and association of SP with CSCs are still not well understood. However, it is now considered that SP is CSC. Studies have shown that OE19 cell line of EAC, which is resistant to 5-fluorouracil, contains a large part of SP and has self-renewal ability and tumorigenesis ability in nude mice (20). Honjo et al. (21) found that ALDH1+ EAC cells sorted out by flow cytometry and fluorescence activation could suspend on the supernatant of serum-free medium to form tumor spheres. Those indicate that ALDH1 is a reliable marker of EAC CSCs. In addition, Wang et al. (22) showed that ALDH+ EAC cells were two to five times more capable of balling in vitro than ALDH-cells, suggesting the expression of ALDH molecules could be used to determine EAC CSCs. Studies have shown that the mRNA and protein expression of integrin α6bri/CD71dim in the sphere-forming cells of various EAC cell lines were more overexpressed than that of the parental cells (23). The suspended sphere-forming EAC cells were significantly stronger than their parental cells in no matter clonogenesis, and tumorigenic ability, or for 5-uracil and cisplatin resistance, because those suspended sphere-forming EAC cells are not common cancer cells, but the latent CSCs, having “stemness”. Under the adaptive conditions, these EAC CSCs will proliferate and differentiate again, causing tumor recurrence and metastasis.
Bile acid, the IL-6/STAT3 signaling pathway and EAC
EAC is an inflammatory cancer with BE as its precancerous lesion. The chronic stimulation of gastroesophageal reflux (the gastroesophageal reflux contains many complex components including gastric acid, bile acid, pepsin, etc.) causes the lower esophagus to be in an inflammatory state, where the local squamous epithelium is gradually replaced by columnar epithelium to form BE. Unfortunately, proton pump inhibitors (PPI) have been widely and frequently used in the treatment of gastric acid inhibition, but in the past few decades, the incidence of EAC in western countries has still increased by 6 times (24). Thus, it suggested that gastric acid is at least not the only key factor inducing EAC. It is currently believed that bile acid is the important reflux that leads to the formation and progression of BE. Bile acids can be classified into free bile acids and bound bile acids according to their structure. Free bile acids include cholic acid, deoxycholic acid (DCA), goose DCA and a small amount of licholic acid. In the rat reflux model, it was found that gastric acid alone could not induce the formation of BE; on the contrary, bile acid alone could induce the formation of BE (25). Shen et al. (13) successfully induced immortalized esophageal squamous cell line Het-1A to reprogram to express intestinal epithelial cell markers CDX2 and MUC2 by utilizing DCA. In addition, compared with patients with gastroesophageal reflux disease and BE without intraepithelial neoplasia, patients with EAC and BE with intraepithelial neoplasia suffered from more severe bile reflux. Therefore, bile acid may be an important environmental factor in the transformation of esophageal squamous cells into BE and carcinogenesis. However, celecoxib, a non-steroidal anti-inflammatory drug, can reduce the expression of CD24, a marker of EAC stem cells, and inhibit the growth of CD24+ EAC in vitro (18). Stimulation of DCA can activate the STAT3 inflammatory pathway and up-regulate the expression of STAT3 (26). Moreover, the upregulation of STAT3 expression could target the expression of stem cell related genes such as OCT4, HIF-1α, and CD44 (27). Upregulation of gene expression in stem cells is an important indicator of stemness acquisition. Meanwhile, DCA stimulation can induce human normal esophageal epithelial cells to transform into BE cells (13). Blocking the stimulation from bile acid or inhibiting inflammation may be a strategy to prevent the formation of CSCs in EAC, and may help to improve the efficacy of radiotherapy and chemotherapy for EAC to prevent tumor recurrence.
IL-6/STAT3 signaling pathway is an inflammatory signaling pathway. Transduction and activation of transcription 3 (STAT3), is a functional protein on the tyrosine phosphorylation signal transducing pathway. Interleukin 6 (IL-6) activates the intracellular downstream Janus kinase (JAK), which further phosphorylates STAT3 to form the active forms of pSTAT3 (Tyr705) and pSTAT3 (Ser727). On one hand, pSTAT3 (Tyr705) is phosphorylated by tyrosine at position 705 of STAT3, and dimerized into the nucleus to target the expression of related downstream genes. On the other hand, pSTAT3 (Ser727) is phosphorylated at Ser727 of STAT3 to regulate gene expression in mitochondria. Studies have shown that cytokine IL-6 can positively feedback to promote the transformation of non-stemness cancer cells into breast cancer CSCs (28) (Figure 1). IL-6 not only mediates the activation of the Notch1 signaling pathway to promote the formation of CSCs (29), but also promotes transformation of cancer cells into CSCs by activating the JAK1-STAT3-OCT4 signal transducing pathway (30). However, the anti-inflammatory substance ginsenoside A can inhibit the activation of IL-6/STAT3 signaling pathway, down-regulate the expression of stem cell genes ALDH1A, SOX2 and OCT4, attenuate the “stemness” to inhibit the ability of breast cancer CSCs to form into globule in vitro (31). In addition, OCT4 was highly expressed in ovarian cancer SP, and the expression of OCT4 was up-regulated in non-SP loaded with OCT4 plasmid, further activating the expression of JAK and STAT, especially JAK1 and STAT6, to improve tumor invasion, anti-apoptosis and tumorigenesis in nude mice (32) (Figure 1). In lung adenocarcinoma, blocking IL-6 can attenuate the formation of lung cancer-like organs from CSCs and increase the sensitivity of lung adenocarcinoma organs to chemotherapeutic drugs (33). In gastric adenocarcinoma, cisplatin induced CSC characteristics are closely related to the activation of IL-6/STAT3 pathway (34). Undoubtedly, IL-6/STAT3 signaling pathway is one of the important pathways to promote the formation of CSCs. During the chronic gastroesophageal reflux, the esophageal epithelial cells are stimulated by bile acid, which can activate inflammatory signaling pathway. High concentration of bile acid can promote the tumorigenesis of digestive tract, because bile acid can induce oxidative stress, DNA damage and mitochondrial damage in cells (35), leading to disease progression, whose specific molecular mechanisms are under study. Studies have shown that bile acid stimulation can induce up-regulated expression of proto-oncogene c-Myc in a variety of EAC cell lines and improve the proliferation capacity of EAC cells (36). Furthermore, DCA stimulation of EAC cells can activate the NF-κB signaling pathway and induce the expression of interleukin IL-8 (37). Other studies have shown that bile acid can activate the IL-6/STAT3 signaling pathway in the esophageal epithelium (38). In addition, Delgado et al. (39) showed that bile acid can promote malignant progression of EAC cells by activating STAT3, up-regulating cyclin D1, cyclin E, c-Myc and Bcl-2, and increasing proliferation and anti-apoptotic ability of EAC. Therefore, bile acid is inextricably linked to the activation of IL-6/STAT3 signaling pathway during the formation and progression of EAC (40). To date, bile acid is known to induce the formation of EAC CSCs. Specifically, whether it mediates this process by activating the IL-6/STAT3 signaling pathway remains to be studied.
Stem cell genes OCT4, KLF4 and the IL-6/STAT3 signaling pathways
The transformation of cancer cells into CSCs is also a process of cell reprogramming. Cell reprogramming is the process by which mature terminal cells are reverted to totipotency or pluripotency, or the formation of pluripotent stem cells, under certain conditions. Under certain conditions, reprogramming of cancer cells changes the malignant degree of cancer cells (Figure 1). In the early stages of reprogramming, the cancer cells become more “stemness” and turn into more malignant CSCs; in the later stage of reprogramming, the pluripotency of cancer cells increases and the degree of malignancy decreases, thus transforming into tumor-IPSCs. The process of cell reprogramming is usually accompanied by over-expression of four Yamanaka transcription factors: KLF4, OCT4, SOX2 and MYC, which are collectively known as OSKM.
Eight dimer transcription factor 4 (OCT4) encoded by POU5F1 genes located on chromosome 6, which is also called POUF51 and OCT3/4. There are related pseudogenes on chromosome 1, 3, 8, 10 and 12, and selective splicing can form different protein subtypes. The standard OCT4 protein sequence contained 360 amino acids with a molecular weight of 38.6 kDa. The POU domain is the main active functional region of OCT4, which is bound to the target gene DNA by the helix-rotation-helix structure. OCT4 is a necessary transcription factor to maintain the multidirectional potential of cells, which is regarded as a stem cell marker and a marker molecule of CSCs. OCT4 and SOX2 are synergistically and directly combined to activate the target gene, and the stem cell factor Nanog is its downstream regulatory gene, whose up-regulation, forming the OCT4-SOX2-Nanog axis is a key cascade of “stemness” (41), in which OCT4 plays an important and core role. Clinically, studies have shown that patients with esophageal squamous cell carcinoma (ESCC) with high expression of OCT4 have poor prognosis (42). Moreover, the abilities of OCT4+ ESCC cells to form tumor sphere and transplanting to form tumors are significantly stronger than that of differentiated mature ESCC cells (43). In addition, OCT3/4+ cell enrichment was also observed in EAC (44). There is an interaction between STAT3 and stem cell factor OCT4 in the regulation between OCT4 and signal pathway. Studies have shown that the use of STAT3 inhibitor WP1066 can inhibit the expression of OCT4 in iPSCs (45). In cervical cancer, the overexpression of STAT3 significantly increased the mRNA and protein expression of OCT4, while the siRNA silencing of STAT3 decreased the expression of OCT4 (46). After transfection with the STAT3 active mutant, pancreatic cancer cells expressed OCT4, the stem cell marker (27). Subsequently, it has been demonstrated that STAT3 can be recruited into the OCT4 promoter region of breast cancer cells (47). However, the relationship between STAT3 and OCT4 in EAC has not been studied.
Kruppel-like factor 4 (KLF4) is a Kruppel like zinc finger protein, which is a member of the DNA binding transcription factor family and is involved in the regulation of various cell functions including proliferation, differentiation, inflammation, apoptosis, cell cycle and embryo development. In previous studies, KLF4 is both a proto-oncogene and a tumor suppressor gene, playing a dual role, which mainly depends on the tissue type and tumor stage of the tumor. In colorectal cancer, gastric cancer, ESCC, lung cancer, prostate cancer and bladder cancer, low expression of KLF4 can promote excessive proliferation and malignant transformation of tumor cells, supporting the role of KLF4 as a tumor suppressor gene. KLF4 is upregulated in primary ductal carcinoma of breast and squamous cell carcinoma of oral cavity, which promotes survival and progression of tumors, reflecting the function of KLF4 as proto-oncogene. Later, KLF4 was found to be involved in the induction of fibroblast transformation into pluripotent induced stem cells (48). As a stem cell gene, KLF4 has attracted much attention, and is involved in maintaining the long-term self-renewal potential of normal stem cells and CSCs. Studies have shown that KLF4 can be activated by bile acid to further increase the transcriptional activity of MUC2 and CDX2, which has a potentially important role in the development of BE (49). Moreover, after inhibiting the Notch1 signaling pathway, esophageal squamous cells can induce the transformation of esophageal squamous cells to BE like metaplasia by up-regulating the expression of KLF4 (15). Therefore, KLF4 is involved in cell reprogramming and can induce the formation of BE. In colon cancer, KLF4 is highly expressed in colon cancer CSCs, and inhibition of KLF4 can reduce the expression of other CSC markers in colon cancer (50). As for the relationship between the STAT3 pathway and KLF4, in embryonic stem cells, the LIF/STAT3 pathway can selectively promote the expression of KLF4 and maintain the pluripotency and self-renewal of embryonic stem cells (51). Studies have shown that the use of STAT3 inhibitor WP1066 can inhibit the expression of KLF4 in iPS cells, suggesting that STAT3 is crucial in maintaining the pluripotency of iPS cells (45). In breast cancer cell lines, the small-molecule Stattic inhibitor of STAT3 significantly reduced the expression level of KLF4 mRNA (52). These studies suggest that KLF4 is a downstream molecule of STAT3, and KLF4 expression is affected by the STAT3 signaling pathway. On the contrary, after treatment with the STAT3 inhibitor STX-0119, an unexpected up-regulation of KLF4 was observed in glioblastoma strains (53). However, the relationship between STAT3 and KLF4 in EAC remains to be studied.
The possible origins of CSCs
It was reported that the CSC population was resistant to chemotherapy. Cancer recurrence is a major obstacle to clinical treatment, in which CSCs may play an important role. One of the explanations for recurrence is that cancer therapy focused too much on destroying tumor cells without eradicating CSCs. Assuming that CSCs behave similarly with normal stem cells, they could act to maintain homeostasis; that is, when the population of tumor cell decreases or disappears through clinical treatment, CSCs would repopulate the tumor and cause recurrence (54). CSCs might be the key drivers of growth and metastasis of the mentioned tumor types and initiator of tumor formation, self-renewal and differentiation.
Although CSCs have been reported in multiple cases of cancer, the source of CSCs has yet to be clear. CSCs might be transformed from adult stem cells, mutations of adult cells, or the activation of proto-oncogenes. On the other hand, they might be transformed by the dedifferentiation of tumor cells who express stem cell related genes indicating their stem cell liked characteristics. If tumor cells express stem cell related genes, it is likely to gain stem cell properties, self-renewal abilities and multiple developing capabilities. Other studies have found that the expression of stem cell markers could be induced in normal tissue cells under certain conditions in which those tissue cells were dedifferentiated into polypotent stem cells and promote tumor formation. According to the present point of view, cancer might be one kind of stem cell disease. The Wnt, Hedgehog and Notch signaling pathways are involved in the expression and regulation of stem cell related genes in tumor cells. In the recent years, studies found that the STAT3 signaling pathway could regulate the expression of stem cell related genes such as Nanog, OCT4, SOX2 and CD44. The tumor cells with high expression of stem cell related genes like Nanog, OCT4, SOX2 and CD44 showed the ability to form balls in vitro and tumors in vivo. That indicated the STAT3 signaling pathway targeted to the expression of stem cell related genes and promoted the formation of tumors. From the therapeutic point of view, it might be an ideal strategy to treat cancer by understanding the biology, focusing on the characteristics in microenvironment and develop specific therapies of CSCs (55).
Conclusions
BE is a precancerous lesion of EAC. As the incidence of BE increases, attention should be paid to the pathogenesis and treatment of EAC. Moreover, CSCs may be the origin of cancer resistance to radiotherapy and chemotherapy, recurrence and metastasis. EAC is an inflammatory tumor with poor prognosis and survival. To explore the relationship between the molecular mechanism of the formation of EAC CSCs and inflammatory signaling pathways may provide strategies for the treatment of EAC.
Acknowledgments
Funding: The work is partly supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.11.12). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hvid-Jensen F, Pedersen L, Drewes AM, et al. Incidence of Adenocarcinoma among Patients with Barrett’s Esophagus. N Engl J Med 2011;365:1375-83. [Crossref] [PubMed]
- Sikkema M, de Jonge PJF, Steyerberg EW, et al. Risk of Esophageal Adenocarcinoma and Mortality in Patients with Barrett’s Esophagus: A Systematic Review and Meta-analysis. Clin Gastroenterol Hepatol 2010;8:235-44. [Crossref] [PubMed]
- Bhat S, Coleman HG, Yousef F, et al. Risk of malignant progression in Barrett’s Esophagus patients: Results from a large population-based study. J Natl Cancer Inst 2011;103:1049-57. [Crossref] [PubMed]
- Dulai GS, Gornbein JK, Kahn L, et al. Preoperative prevalence of Barrett’s esophagus in esophageal adenocarcinoma: A systematic review. Gastroenterology 2002;122:26-33. [Crossref] [PubMed]
- Ruol A, Parenti A, Zaninotto G, et al. Intestinal metaplasia is the probable common precursor of adenocarcinoma in Barrett esophagus and adenocarcinoma of the gastric cardia. Cancer 2000;88:2520-8. [Crossref] [PubMed]
- Shapiro J, van Lanschot JJB, Hulshof MCCM, et al. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): Long-term results of a randomised controlled trial. Lancet Oncol 2015;16:1090-8. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Donadelli M, Dando I, Pozza ED, et al. Mitochondrial uncoupling protein 2 and pancreatic cancer: A new potential target therapy. World J Gastroenterol 2015;21:3232-8. [Crossref] [PubMed]
- Darlavoix T, Seelentag W, Yan P, et al. Altered expression of CD44 and DKK1 in the progression of Barrett’s esophagus to esophageal adenocarcinoma. Virchows Arch 2009;454:629-37. [Crossref] [PubMed]
- Minacapelli CD, Bajpai M, Geng X, et al. Barrett’s metaplasia develops from cellular reprograming of esophageal squamous epithelium due to gastroesophageal reflux. Am J Physiol - Gastrointest Liver Physiol 2017;312:G615-22. [Crossref] [PubMed]
- Aikou S, Aida J, Takubo K, et al. Columnar metaplasia in a surgical mouse model of gastro-esophageal reflux disease is not derived from bone marrow-derived cell. Cancer Sci 2013;104:1154-61. [Crossref] [PubMed]
- Barbera M, Fitzgerald RC. Cellular origin of Barrett’s metaplasia and oesophageal stem cells. Biochem Soc Trans 2010;38:370-3. [Crossref] [PubMed]
- Shen C, Zhang H, Wang P, et al. Deoxycholic acid (DCA) confers an intestinal phenotype on esophageal squamous epithelium via induction of the stemness-associated reprogramming factors OCT4 and SOX2. Cell Cycle 2016;15:1439-49. [Crossref] [PubMed]
- Yan W, Zhang H, Li J, et al. BMP4 promotes a phenotype change of an esophageal squamous epithelium via up-regulation of KLF4. Exp Mol Pathol 2016;101:259-66. [Crossref] [PubMed]
- Vega ME, Giroux V, Natsuizaka M, et al. Inhibition of notch signaling enhances transdifferentiation of the esophageal squamous epithelium towards a Barrett’s-like metaplasia via KLF4. Cell Cycle 2014;13:3857-66. [Crossref] [PubMed]
- Yu WY, Slack JMW, Tosh D. Conversion of columnar to stratified squamous epithelium in the developing mouse oesophagus. Dev Biol 2005;284:157-70. [Crossref] [PubMed]
- Smit JK, Faber H, Niemantsverdriet M, et al. Prediction of response to radiotherapy in the treatment of esophageal cancer using stem cell markers. Radiother Oncol 2013;107:434-41. [Crossref] [PubMed]
- Jiménez P, Chueca E, Arruebo M, et al. CD24 Expression Is Increased in 5-Fluorouracil-Treated Esophageal Adenocarcinoma Cells. Front Pharmacol 2017;8:321. [Crossref] [PubMed]
- Sun X, Martin GCG, Zheng Q, et al. Drug-induced expression of EpCAM contributes to therapy resistance in esophageal adenocarcinoma. Cell Oncol (Dordr) 2018;41:651-62. [Crossref] [PubMed]
- Zhao Y, Bao Q, Schwarz B, et al. Stem Cell-Like Side Populations in Esophageal Cancer: A Source of Chemotherapy Resistance and Metastases. Stem Cells Dev 2014;23:180-92. [Crossref] [PubMed]
- Honjo S, Ajani JA, Scott AW, et al. Metformin sensitizes chemotherapy by targeting cancer stem cells and the mTOR pathway in esophageal cancer. Int J Oncol 2014;45:567-74. [Crossref] [PubMed]
- Wang Z, Da Silva TG, Jin K, et al. Notch signaling drives stemness and tumorigenicity of esophageal adenocarcinoma. Cancer Res 2014;74:6364-74. [Crossref] [PubMed]
- Zhao R, Quaroni L, Casson AG. Identification and characterization of stemlike cells in human esophageal adenocarcinoma and normal epithelial cell lines. J Thorac Cardiovasc Surg 2012;144:1192-9. [Crossref] [PubMed]
- Hur C, Miller M, Kong CY, et al. Trends in esophageal adenocarcinoma incidence and mortality. Cancer 2013;119:1149-58. [Crossref] [PubMed]
- Sun D, Wang X, Gai Z, et al. Bile acids but not acidic acids induce Barrett’s esophagus. Int J Clin Exp Pathol 2015;8:1384-92. [PubMed]
- Zhang HY, Zhang Q, Zhang X, et al. Cancer-related inflammation and Barrett’s carcinogenesis: interleukin-6 and STAT3 mediate apoptotic resistance in transformed Barrett’s cells. Am J Physiol Gastrointest Liver Physiol 2011;300:G454-60. [Crossref] [PubMed]
- Tyagi N, Marimuthu S, Bhardwaj A, et al. P-21 activated kinase 4 (PAK4) maintains stem cell-like phenotypes in pancreatic cancer cells through activation of STAT3 signaling. Cancer Lett 2016;370:260-7. [Crossref] [PubMed]
- Iliopoulos D, Hirsch HA, Wang G, et al. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci 2011;108:1397-402. [Crossref] [PubMed]
- Hsu EC, Kulp SK, Huang HL, et al. Function of integrin-linked kinase in modulating the stemness of il-6-abundant breast cancer cells by regulating γ-secretase-mediated notch1 activation in caveolae. Neoplasia 2015;17:497-508. [Crossref] [PubMed]
- Kim SY, Kang JW, Song X, et al. Role of the IL-6-JAK1-STAT3-Oct-4 pathway in the conversion of non-stem cancer cells into cancer stem-like cells. Cell Signal 2013;25:961-9. [Crossref] [PubMed]
- Liu C, Dong L, Sun Z, et al. Esculentoside A suppresses breast cancer stem cell growth through stemness attenuation and apoptosis induction by blocking IL-6/STAT3 signaling pathway. Phytother Res 2018;32:2299-311. [Crossref] [PubMed]
- Ruan Z, Yang X, Cheng W. OCT4 accelerates tumorigenesis through activating JAK/STAT signaling in ovarian cancer side population cells. Cancer Manag Res 2018;11:389-99. [Crossref] [PubMed]
- Ogawa H, Koyanagi-Aoi M, Otani K, et al. Interleukin-6 blockade attenuates lung cancer tissue construction integrated by cancer stem cells. Sci Rep 2017;7:12317. [Crossref] [PubMed]
- Wang X, Li Y, Dai Y, et al. Sulforaphane improves chemotherapy efficacy by targeting cancer stem cell-like properties via the miR-124/IL-6R/STAT3 axis. Sci Rep 2016;6:36796. [Crossref] [PubMed]
- Bernstein H, Bernstein C, Payne CM, et al. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res 2005;589:47-65. [Crossref] [PubMed]
- Tselepis C, Morris CD, Wakelin D, et al. Upregulation of the oncogene c-myc in Barrett’s adenocarcinoma: induction of c-myc by acidified bile acid in vitro. Gut 2003;52:174-80. [Crossref] [PubMed]
- Jenkins GJ, Harries Doak SH, Wilmes Griffiths AP, et al. The bile acid deoxycholic acid (DCA) at neutral pH activates NF-κB and induces IL-8 expression in oesophageal cells in vitro. Carcinogenesis 2004;25:317-23. [Crossref] [PubMed]
- Goldman A, Chen HD, Rosely HB, et al. Characterization of squamous esophageal cells resistant to bile acids at acidic pH: implication for Barrett’s esophagus pathogenesis. Am J Physiol Gastrointest Liver Physiol 2011;300:G292-302. [Crossref] [PubMed]
- Delgado JS, Mustafi R, Yee J, et al. Sorafenib triggers antiproliferative and pro-apoptotic signals in human esophageal adenocarcinoma cells. Dig Dis Sci 2008;53:3055-64. [Crossref] [PubMed]
- Dvorak K, Chavarria M, Payne CMK, et al. Activation of the interleukin 6/STAT3 antiapoptotic pathway in esophageal cells by bile acids and low pH: Relevance to Barrett’s esophagus. Clin Cancer Res 2007;13:5305-13. [Crossref] [PubMed]
- Boyer LA, Lee TI, Cole MF, et al. Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells. Cell 2005;122:947-56. [Crossref] [PubMed]
- He W, Li K, Wang F, et al. Expression of OCT4 in human esophageal squamous cell carcinoma is significantly associated with poorer prognosis. World J Gastroenterol 2012;18:712-9. [Crossref] [PubMed]
- Zhou X, Huang GR, Hu P. Over-expression of Oct4 in human esophageal squamous cell carcinoma. Mol Cells 2011;32:39-45. [Crossref] [PubMed]
- Mendelson J, Song S, Li Y, et al. Dysfunctional TGF-β signaling with constitutively active Notch signaling in Barrett’s esophageal adenocarcinoma. Cancer 2011;117:3691-702. [Crossref] [PubMed]
- Kuan II, Liang KH, Wang YP, et al. EpEX/EpCAM and Oct4 or Klf4 alone are sufficient to generate induced pluripotent stem cells through STAT3 and HIF2α. Sci Rep 2017;7:41852. [Crossref] [PubMed]
- Wang H, Deng J, Ren HY, et al. STAT3 influences the characteristics of stem cells in cervical carcinoma. Oncol Lett 2017;14:2131-6. [Crossref] [PubMed]
- Sengupta S, Nagalingam A, Muniraj N, et al. Activation of tumor suppressor LKB1 by honokiol abrogates cancer stem-like phenotype in breast cancer via inhibition of oncogenic Stat3. Oncogene 2017;36:5709-21. [Crossref] [PubMed]
- Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006;126:663-76. [Crossref] [PubMed]
- Kazumori H, Ishihara S, Takahashi Y, et al. Roles of Krüppel-like factor 4 in esophageal epithelial cells in Barrett’s epithelium development. Gut 2011;60:608-17. [Crossref] [PubMed]
- Leng Z, Tao K, Xia Q, et al. Krüppel-Like Factor 4 Acts as an Oncogene in Colon Cancer Stem Cell-Enriched Spheroid Cells. PLoS One 2013;8:e56082. [Crossref] [PubMed]
- Hall J, Guo G, Wray J, et al. Oct4 and LIF/Stat3 Additively Induce Krüppel Factors to Sustain Embryonic Stem Cell Self-Renewal. Cell Stem Cell 2009;5:597-609. [Crossref] [PubMed]
- Yang CM, Chiba T, Groner B, et al. Expression of reprogramming factors in breast cancer cell lines and the regulation by activated Stat3. Horm Mol Biol Clin Investig 2012;10:241-8. [Crossref] [PubMed]
- Ashizawa T, Miyata H, Lizuka A, et al. Effect of the STAT3 inhibitor STX-0119 on the proliferation of cancer stem-like cells derived from recurrent glioblastoma. Int J Oncol 2013;43:219-27. [Crossref] [PubMed]
- Abernathy K, Burke J. Modeling the Treatment of Glioblastoma Multiforme and Cancer Stem Cells with Ordinary Differential Equations. Comput Math Methods Med 2016;2016:1239861. [Crossref] [PubMed]
- Gao XM, Zhang R, Dong QZ, et al. Properties and feasibility of using cancer stem cells in clinical cancer treatment. Cancer Biol Med 2016;13:489-95. [Crossref] [PubMed]


