
Functional analysis of long-chain non-coding RNA in oral squamous cell carcinoma
Introduction
Non-coding RNAs (ncRNAs) include long-chain non-coding RNA (lncRNA), circRNA and microRNA (1). LncRNAs are a group of RNA molecules with transcripts over 200 nt in length and cannot encode proteins. LncRNAs were originally regarded as the “noise” in the genomic transcription and the by-products of RNA polymerase II medicated transcription because they have no biological functions (2). However, recent studies have shown that lncRNAs are involved in the regulation of many important processes such as X chromosome silencing, genomic imprinting, chromatin modification, transcriptional activation, transcriptional interference and intra-nuclear transport (3-6). These regulatory effects of lncRNAs have attracted wide attention, and great progresses have been achieved in the relationship between lncRNAs and tumors. However, little is known about the specific mechanism underlying the lncRNA mediated regulation, and the roles of lncRNAs in the invasion and metastasis of oral squamous cell carcinoma (OSCC) are still unclear.
In the present study, high-throughput microarray was employed to investigate the expression profile of lncRNA in the OSCC tissues. The differentially expressed lncRNAs and mRNA in the OSCCs as compared with adjacent normal tissues as well as their biological functions were analyzed. Our findings may provide experimental evidence for exploring the mechanism underlying the occurrence and development of OSCC and also present new biomarkers and therapeutic targets for OSCC.
Methods
Sample collection and processing
From January 2015 to January 2018, 30 patients with OSCC who underwent first operation in the Department of Oral and Maxillofacial Surgery, Hangzhou First Hospital, Zhejiang University were recruited into present study. There were 17 males and 13 females, and the mean age was 62.4 years old (range: 30–75 years old). This study was approved by the Ethics Committee of Hangzhou First Hospital, Zhejiang University (2019-015-01). All the patients did not receive chemotherapy, radiotherapy and immunotherapy before surgery. Informed consent was obtained from each patient or their relatives before study.
The cryotube was numbered and sterilized. Tissues included tumor tissues and adjacent normal tissues. The tumor tissues were collected out of the necrosis region and the adjacent normal tissues were collected 3 cm away from the tumor (about 0.5 cm × 0.5 cm × 0.5 cm). These tissues were divided into two parts: one was immediately placed into RNAlater solution tissue preservation (v:v, 1:10). Subsequently, the tissues were transferred into a cryotube and stored in liquid nitrogen and then at −80 °C. The remaining tissues were fixed in formalin for further pathological and immunohistochemical examinations. The pathological diagnosis and histological classification were made by two experienced pathologists in the Hangzhou First Hospital, Zhejiang University in accordance with the WHO diagnostic criteria. Tissues were collected from 5 patients and processed for expression profiling of lncRNA/mRNA, and the remaining 25 pairs of samples were processed for real-time quantitative PCR (QuantitatiVe ReaI-time PoIymerase Chain Reactjon, qRT-PCR).
Microarray assay of lncRNA
The differentially expressed lncRNA and mRNAs were examined in 5 pairs of tissues with the 4×180 K lncRNA + mRNA Human Gene Expression Microarray V4.0 (CapitalBio Technology, Beijing, China). The lncRNA data of this microarray are from NCIB Refseq, UCSC Known Gene6.0, Gencode v13, RNAdb, NRED and LincRNAs, and mRNA data were from the collaborative consensus coding sequence (CCDS) project. Each microarray includes 4 isolated matrices and each matrix includes about 1.7×105 probes. It can detect the expression of both lncRNA and mRNA. The microarray was developed by the Beijing Bo’ao Jingdian Biotechnology Co., Ltd. The microarray was 4×180 K and each microarray includes 40,916 lncRNAs and 34,235 mRNAs.
Pre-processing of expression data
Total RNA was extracted for in vitro amplification and fluorescence labeling with Jingxin® Biochip general labeling kit. Total RNA, T7 Oligo(dT)Primer, T7 specific primers [can bind to mRNA containing poly(A) and RNA without poly(A) except for rRNA] and First Strand Enzyme Mix were used for the reverse transcription of First strand cDNA. The RNA in DNA-RNA mixture was transcribed into Second Strand cDNA with Second Strand Enzyme Mix. Then, the Second Strand cDNA as a template and T7 Enzyme Mix were used for the synthesis of cRNA which was subsequently purified with RNA purification column to remove the salt and enzyme. The cRNA was then subjected to quantification and quality control. After reverse transcription, the cDNA was purified, retrieved and quantified. The products after cDNA reverse transcription as templates, Random Primer and Klenow Fragment enzyme were used to synthesize complementary cDNA, which was then labeled with fluorescent dNTP, purified and quantified. The fluorescent DNAs were purified with Nucleospin® Extract II kit, and volume after elution was 30 µL. The cy3-dCTP labeled products after elution were concentrated in the vacuum or mixed with water to a volume of 27.5 µL. Hybridization was done at 45 °C over night.
Microarray rinsing and scanning
After detection, the microarray was rinsed in the Slide Washer8 rinsing machine and then dried. The microarray was scanned with the Agilent scanner (G2565CA), and then photographed.
Microarray analysis
Agilent Feature Extraction (v10.7) software (Santa Clara, CA, USA) was used to analyze the hybridization map and extract the data. The data normalization and difference analysis were performed with Agilent GeneSpring software. The raw data were standardized, and the high-quality probes were screened for further analysis. The two groups of sample data underwent a t-test analysis to obtain the corrected P-values and Fold Change values. The criteria for the differentially expressed genes that changed were more than twofold, and the differences were considered to be statistically significant at P<0.05. Cluster 3.0 software (Michiel de Hoon, Human Genome Center, University of Tokyo) was also used for the cluster analysis and graphical display. The differentially expressed genes were obtained by comparing two groups. The differentially expressed mRNAs were then subjected to GO & Pathway and Disease analyses.
Functional analysis of mRNA and screening of key lncRNA
The differentially expressed mRNAs were subjected to GO analysis, Pathway analysis and disease enrichment analysis [GO: Gene Ontology including cellular component (CC); molecular function: MF; biological process: BP. Pathway including KEGG, PID, BioCarta, Reactome, Panther and BioCyc; Disease including OMIM, KEGG DISEASE, FunDO, GAD, NHGRI and Disease]. Based on the items closely related to the tumor and the co-expression network of the lncRNAs and mRNAs, key lncRNAs related to OSCC were screened.
Statistical analysis
Agilent GeneSpring GX v12.1 was used for the analysis of differentially expressed lncRNA and mRNA. Comparisons between groups were done with t-test. The criteria for the differentially expressed genes that changed were more than twofold, and the differences were considered to be statistically significant at P<0.05.
Results
Quality control of total RNA
Results showed that the OD260/OD280 of RNA ranged from 1.8 to 2.1, and the OD260/OD230 was 2.2–2.4. This suggests the extracted RNA has a high purity. Further agarose gel electrophoresis showed the 5S, 18S and 28S bands of RNA were complete, and the brightness of the 28S band was twice that of the 18S band. This indicates the integrity of the extracted RNA was good for the following experiment.
Data normalization and principal component analysis (PCA)
After hybridization of lncRNA, the map data were pre-processed with feature extraction. Data normalization and QC analysis showed the consistent trends in the genes and showed high similarity (Figures 1 and 2).
Expression profiles of lncRNA and mRNA
The signals from high-throughput assay were scanned and analyzed, and the lncRNAs and mRNAs with fold change of ≥2.0 and P<0.05 were defined as differentially expressed lncRNA and mRNA, respectively. Subsequently, Cluster 3.0 software was used for hierarchical clustering analysis, and the data were visualized with the treeview software to identify the differences in the differentially expressed lncRNA and mRNA between tumor tissues and adjacent normal tissues. As compared with adjacent normal tissues as controls, a total of 3,022 differentially expressed lncRNAs and 4,364 differentially expressed mRNAs were found in the tumor tissues. A total of 1,453 lncRNAs showed up-regulated expression and 1,569 lncRNAs exhibited down-regulated in the tumor tissues; 2,851 mRNAs showed increased expression and 1,513 mRNAs had reduced expression. Among them, there were 853 antisense lncRNAs, 75 sense lncRNAs, 118 bidirectional and intragenic lncRNAs, 1,463 intergenic lncRNA and 512 other lncRNAs.
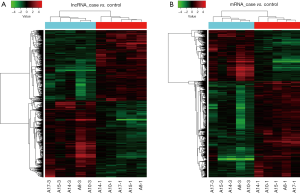
Heat map and hierarchical clustering
Cluster_*RNA.txt data were loaded with the Cluster 3.0 software. The signal values of genes after data normalization were subjected to individualized clustering analysis. Then, the tree view software was used for visualization adjustment and graphic output. The red in the heat map indicates relatively high expression, and the blue indicates relatively low expression. According to the differences in expression of lncRNAs and mRNAs between groups, hierarchical clustering analysis was done to cluster samples with similar expression level, and then the relationship between different samples was further analyzed (Figure 3).
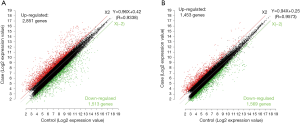
Scatter plots
The scatter plot was used to intuitively analyze the distribution variation of data from microarray assay in the two groups. The X-axis and the Y-axis of the scatter plot are the signal values after normalization, and the green line indicates the fold change. The plots above and below the green line represented the lncRNAs and mRNAs with the fold change of expression ≥2.0 in the OSCC tissues as compared with adjacent normal tissues (Figure 4).
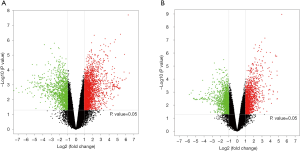
Volcanoplot
The volcanoplot was determined with the P value and FC value obtained by the difference analysis, aiming to show the significant difference between two groups (Figure 5).
Bioinformatics analysis of differentially expressed mRNAs
The differentially expressed mRNAs were subjected to GO, Pathway, and disease enrichment annotation (GO: Gene Ontology, including cellular component, CC; molecular function, MF; biological process, BP). The GO database is an international standard classification system for gene function and applicable to various species. It can define and describe genes and proteins. In the GO analysis, genes are classified according to the Cellular component, Molecular Function and Biological process. Pathways include KEGG, PID, BioCarta, Reactome, Panther and BioCyc; Diseases contain OMIM, KEGG DISEASE, FunDO, GAD, NHGRI and Disease.
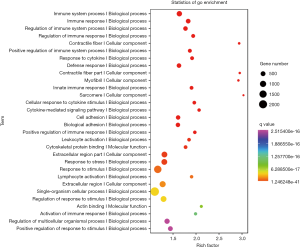
Gene ontology analysis of differentially expressed mRNAs
The differentially expressed mRNAs were subjected to GO enrichment analysis and the distribution of differentially expressed mRNAs in GO was further investigated, aiming to elucidate the function of these mRNAs. First, all the differentially mutated genes were mapped to various terms of the GO database (http://www.geneontology.org/), and then the number of genes in each term was calculated. A hypergeometric test was employed to identify the significantly enriched items of mutated genes in the GO at the whole genome background. After multiple test adjustment, a GO term with q value ≤0.05 was defined as the GO term with significant enrichment in the mutated genes. By using the GO database, a clustering analysis of gene functions was done for all these differentially expressed mRNAs in tumor tissues. Figure 6 shows the results of GO enrichment analysis (top 30 items).
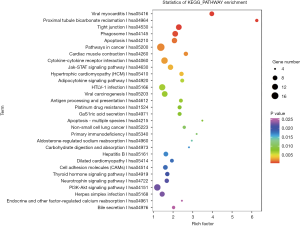
Pathway annotation and enrichment analysis of differentially expressed mRNAs
The differentially expressed mRNAs were subjected to pathway annotation and enrichment. The related databases included KEGG PATHWAY, PID, BioCarta, Reactome, BioCyc and PANTHER. The significant enrichment can identify the most important biochemical metabolic pathways and signal transduction pathways related to differentially expressed genes. After multiple test for adjustment, the pathway with P value ≤0.05 was defined as the pathway with significant enrichment related to the mutated genes (Figure 7).
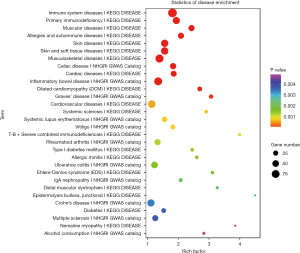
Disease annotation analysis of differentially expressed genes
The differentially expressed mRNAs screened were searched in the human disease database, aiming to identify the differentially expressed mRNAs in the relevant database. It mainly included five human disease databases, namely OMIM, KEGG DISEASE, FunDO, GAD and NHGRI GWAS Catalog. After multiple test adjustment, pathway with P value ≤0.05 was defined as the disease with significant enrichment in the mutated genes (Figure 8).
Screening of differentially expressed lncRNAs in the OSCC tissues
The 4,364 differentially expressed mRNAs were subjected to GO (Gene Oncology, GO), Pathway, and disease analysis and items closely related to tumors were employed for the screening of differentially expressed mRNAs. Based on Disease:Cancers, Head and neck cancers, GO:cell adhesion, cell adhesion, Pathway: tight junction and Pathways in cancer, and Jak-STAT signaling pathway, a total of 130 major differentially expressed mRNAs were identified.
Based on the co-expression of the lncRNAs and mRNAs (correlation 40.99 or correlation −0.99 and P value <0.05), there were differentially expressed mRNAs with co-expression with lncRNA. Moreover, the intersection analysis between 130 gene symbols and 73 gene symbols showed a total of nine miRNAs, and the number of targeted lncRNA was 11, which served as the differentially expressed lncRNA (Table 1). Based on the limitations of items and the lncRNA/mRNA co-expression, 9 differentially expressed mRNAs and 11 differentially expressed lncRNA were identified. Of 11 lncRNA, 5 were up-regulated [uc.472+ (FC =2.454442076), ENSG00000267458.1 (FC =2.560549258), ENSG00000257922.1 (FC =2.889767092), ENSG00000258908.1 (FC =4.626702884), HIX0000266 (FC =4.896329302)] and 6 were down-regulated [HIX0011161 (FC =2.722727955), HIX0026549 (FC =3.9141725), AK055408 (FC =4.345569414), ENSG00000258444.1 (FC =9.951425458), ENSG00000214970.3 (FC =47.14573143), ENSG00000257514.1 (FC =55.11227667)].
Table 1
| lncRNA | P | FC (abs) | Regulation | Alias |
|---|---|---|---|---|
| uc.472+ | 0.014124248 | 2.454442076 | Up | UCRs_462_202 |
| ENSG00000267458.1 | 0.007448052 | 2.560549258 | Up | ENST00000589120.1 |
| ENSG00000257922.1 | 0.000171036 | 2.889767092 | Up | ENST00000549285.1 |
| ENSG00000258908.1 | 0.013095483 | 4.626702884 | Up | ENST00000554678.1 |
| HIX0000266 | 0.003438903 | 4.896329302 | Up | H-InvDB_2878_218 |
| HIX0011161 | 0.002920305 | 2.722727955 | Down | H-InvDB_506_629 |
| HIX0026549 | 0.006478557 | 3.9141725 | Down | H-InvDB_1815_323 |
| AK055408 | 0.001038765 | 4.345569414 | Down | uc001unn.1 |
| ENSG00000258444.1 | 0.001503996 | 9.951425458 | Down | ENST00000557368.1 |
| ENSG00000214970.3 | 0.001174612 | 47.14573143 | Down | ENST00000581304.1 |
| ENSG00000257514.1 | 0.001524591 | 55.11227667 | Down | ENST00000547027.1 |
LncRNA, long-chain non-coding RNA.
Discussion
OSCC is the most common malignant tumor of the mucosa epithelium in the oral and maxillofacial region and accounts for more than 80% of oral cancers. OSCC is common in the elderly and has been one of the top ten cancers worldwide. It is the third most common cancer in the developing countries and the sixth common cancer worldwide. Moreover, the incidence of OSCC is increasing over year, and about 500,000 new OSCC cases are diagnosed in each year (7,8). Although great effort has been paid by the governments, medical institutions, universities, pharmaceutical companies and charitable foundations to the investigations about the pathogenesis, diagnosis and treatment of OSCC and great progress has been achieved in the chemotherapy, biotherapy, and gene therapy of OSCC, the long-term efficacy of these treatments is still unsatisfactory (9-11).
OSCC has become a serious threat to human health and life. In the past two decades, although great progress has been achieved in the detection techniques, surgical treatment, radiotherapy, chemotherapy, biotherapy and gene therapy of OSCC, OSCC is often diagnosed at advanced stage in a majority of patients, and radical surgery is still the main treatment for OSCC. Moreover, the long term efficacy of surgery is still unsatisfactory, the prognosis is still poor and the 5-year survival is at a low level in patients with recurrence and/or metastasis. The survival rate is no higher than 50% in patients with unilateral lymph node metastasis and no higher than 25% in patients with bilateral lymph node metastasis, and the overall survival rate is lower than 50% (12-14).
Thus, to improve the therapeutic efficacy, reduce the postoperative recurrence and metastasis, and effectively prolong the survival time of patients have become important issues in the management of OSCC (15-18) Favorable efficacy has been achieved in the targeted therapy of several malignancies such as breast cancer and lung cancer (19-23). For some patients with advanced OSCC who have poor responses to available therapies, targeted therapy may become a very important auxiliary treatment. In fact, the targeted therapy of OSCC has attracted much attention. Unfortunately, a variety of studies have been conducted to investigate the mechanism underlying the occurrence, development, invasion and metastasis of OSCC, the effective targets for the targeted therapy for OSCC are still unclear (24,25).
Previous studies have shown that abnormal gene expression and regulation may be important cause and basis of the occurrence and development of malignant tumors (26,27). With the deepening of genomic researches, many new important molecules and mechanisms are identified. Especially, recent studies have shown that the lncRNAs may play important roles in the occurrence and development of OSCC and may become potential targets in the targeted therapy of OSCC.
Studies have shown that the genes encoding proteins account for less than 3% of the human genome, and more than 80% of the genome are the RNA transcripts that do not encode proteins. Such transcripts are also known as ncRNAs. According to the size, ncRNAs can be divided into lncRNA and short-chain ncRNAs. LncRNAs refer to RNAs with more than 200 nucleotides and unable to encode proteins. Studies have revealed that lncRNAs play crucial roles in the regulation of gene expression at the epigenetic, transcriptional and post-transcriptional levels (28).
Recent studies have shown that lncRNAs are involved in many important regulatory processes such as X-chromosome silencing, genomic imprinting, chromatin modification, transcriptional activation, transcriptional interference, and intranuclear transport. They can act as baits, activators, guides or scaffolds among interacting proteins (29). In addition to these transcriptional and epigenetic regulations, lncRNAs have also been found to play an important role in the post-transcriptional regulation, such as mRNA splicing, mRNA editing, and storage of small ncRNAs. In addition, lncRNAs can interact with a variety of biological macromolecules, such as: chromosomes, mRNA, hacRNA and proteins to exert biological effects. For example, lncRNA-BCl, lncRNA-P21 and lncRNA-UCHl can bind to receptor proteins to reduce the stability of proteins, which inhibits their activity (30-32). lncRNA-HOTAIR is able to bind AR to stabilize AR protein, thereby increasing its activity (33). Further studies have confirmed that lncRNAs play important roles in the occurrence and development of tumors, and the abnormal expression of genes regulated by lncRNAs may cause serious pathological changes, which is one of the important causes of diseases (34). There is evidence showing that lncRNA can affect the proliferation of tumor cells and these cells may escape from the cytokine induced inhibition, which results in infinite cell proliferation, ultimately inducing angiogenesis and avoidance of cell death, In addition, abnormal expression of lncRNAs is closely related to the tumorigenesis, diagnosis, metastasis and prognosis, which provide new directions for researches about targeted therapy of cancers. However, the specific mechanisms underlying the regulation of lncRNAs are largely unclear, and more studies are needed. In particular, few studies have been conducted to investigate the mechanism underlying the regulatory role of lncRNAs in the OSCC.
In the present study, differentially expressed lncRNAs and mRNAs in OSCC tissues and adjacent normal tissues were screened by lncRNA/mRNA high-throughput microarray technique. A total of 3,022 differentially expressed lncRNAs and 4,364 differentially expressed mRNAs were found in the OSCC tissues as compared with adjacent normal tissues as controls. Then, KOBAS software was used to perform bioinformatics analysis on GO, Pathway, and disease (human species only) annotations. Disease: Cancers, Head and neck cancers; GO: cell adhesion, cell proliferation, regulation of cell proliferation, regulation of cell adhesion; Pathway: tight junction; Pathways in cancer, Jak-STAT signaling pathway as well as terms related to the occurrence and development of tumors were employed to limit the range of differentially expressed mRNA in the OSCC tissues. It is well known that lncRNAs can interact with adjacent genes to regulate their expression, or indirectly affect the expression of distant genes through the miRNAs. Based on the lncRNA-mRNA co-expression (correlation >0.99 or correlation <−0.99, and P value <0.05), the cis-prediction involved the lncRNA-mRNA pair located within 10 kB in the genome, and the trans-prediction involved the sequence similarity lncRNA-mRNA pairs after comparing the lncRNA and mRNA sequences (3'UTR) using the BLAST tool. Then, the lncRNA/mRNA co-expression network was constructed.
Cancer, head and neck cancer, cell adhesion, cell proliferation, regulation of cell proliferation, regulation of cell adhesion, tight junctions, cancer pathways and JAK-STAT signaling pathways are prerequisites for screening critical lncRNAs in the present study. That is, the biological functions of these identified lncRNAs should be related to the above items. Therefore, we speculate that the information in this study may provide important reference for further investigations on the pathogenesis of OSCC, and contribute to the identification of therapeutic targets and biomarkers of OCSS.
Acknowledgments
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.67). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Hangzhou First Hospital, Zhejiang University (2019-015-01). Informed consent was obtained from each patient or their relatives before study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yang JX, Rastetter RH, Wilhelm D. Non-coding RNAs: An Introduction. Adv Exp Med Biol 2016;886:13-32. [Crossref] [PubMed]
- Jarroux J, Morillon A, Pinskaya M. History, Discovery, and Classification of lncRNAs. Adv Exp Med Biol 2017;1008:1-46. [Crossref] [PubMed]
- Lu Z, Carter AC, Chang HY. Mechanistic insights in X-chromosome inactivation. Philos Trans R Soc Lond B Biol Sci 2017;372. [PubMed]
- Kanduri C. Long noncoding RNAs: Lessons from genomic imprinting. Biochim Biophys Acta 2016;1859:102-11. [Crossref] [PubMed]
- Han P, Chang CP. Long non-coding RNA and chromatin remodeling. RNA Biol 2015;12:1094-8. [Crossref] [PubMed]
- Liu SJ, Horlbeck MA, Cho SW, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 2017;355. [PubMed]
- Tandon P, Dadhich A, Saluja H, et al. The prevalence of squamous cell carcinoma in different sites of oral cavity at our Rural Health Care Centre in Loni, Maharashtra - a retrospective 10-year study. Contemp Oncol (Pozn) 2017;21:178-83. [Crossref] [PubMed]
- Gudiseva S, Santosh ABR, Chitturi R, et al. The role of mast cells in oral squamous cell carcinoma. Contemp Oncol (Pozn) 2017;21:21-9. [Crossref] [PubMed]
- Baillie R, Tan ST, Itinteang T. Cancer Stem Cells in Oral Cavity Squamous Cell Carcinoma: A Review. Front Oncol 2017;7:112. [Crossref] [PubMed]
- Wu YL, Li HY, Zhao XP, et al. Mesenchymal stem cell-derived CCN2 promotes the proliferation, migration and invasion of human tongue squamous cell carcinoma cells. Cancer Sci 2017;108:897-909. [Crossref] [PubMed]
- Li H, Wang H, Sun Z, et al. The clinical and prognostic value of polo-like kinase 1 in lung squamous cell carcinoma patients: immunohistochemical analysis. Biosci Rep 2017; [Epub ahead of print]. [Crossref] [PubMed]
- Lin XJ, He CL, Sun T, et al. hsa-miR-485-5p reverses epithelial to mesenchymal transition and promotes cisplatin-induced cell death by targeting PAK1 in oral tongue squamous cell carcinoma. Int J Mol Med 2017;40:83-9. [Crossref] [PubMed]
- Hu Q, Wu T, Chen X, et al. The poor outcome of second primary oral squamous cell carcinoma is attributed to Bmi1 upregulation. Cancer Med 2018;7:1056-69. [Crossref] [PubMed]
- Moriwaki K, Ayani Y, Kuwabara H, et al. TRKB tyrosine kinase receptor is a potential therapeutic target for poorly differentiated oral squamous cell carcinoma. Oncotarget 2018;9:25225-43. [Crossref] [PubMed]
- Mitra AK, Agrahari V, Mandal A, et al. Novel delivery approaches for cancer therapeutics. J Control Release 2015;219:248-68. [Crossref] [PubMed]
- Zielonka J, Joseph J, Sikora A, et al. Mitochondria-Targeted Triphenylphosphonium-Based Compounds: Syntheses, Mechanisms of Action, and Therapeutic and Diagnostic Applications. Chem Rev 2017;117:10043-120. [Crossref] [PubMed]
- Marks EI, Yee NS. Molecular genetics and targeted therapeutics in biliary tract carcinoma. World J Gastroenterol 2016;22:1335-47. [Crossref] [PubMed]
- Au JL, Yeung BZ, Wientjes MG, et al. Delivery of cancer therapeutics to extracellular and intracellular targets: Determinants, barriers, challenges and opportunities. Adv Drug Deliv Rev 2016;97:280-301. [Crossref] [PubMed]
- Shull JD, Dennison KL, Chack AC, et al. Rat models of 17beta-estradiol-induced mammary cancer reveal novel insights into breast cancer etiology and prevention. Physiol Genomics 2018;50:215-34. [Crossref] [PubMed]
- Wehde BL, Radler PD, Shrestha H, et al. Janus Kinase 1 Plays a Critical Role in Mammary Cancer Progression. Cell Rep 2018;25:2192-207.e5. [Crossref] [PubMed]
- Blandin Knight S, Crosbie PA, Balata H, et al. Progress and prospects of early detection in lung cancer. Open Biol 2017; [Crossref] [PubMed]
- Morgensztern D, Campo MJ, Dahlberg SE, et al. Molecularly targeted therapies in non-small-cell lung cancer annual update 2014. J Thorac Oncol 2015;10:S1-63. [Crossref] [PubMed]
- Ji X, Bosse Y, Landi MT, et al. Identification of susceptibility pathways for the role of chromosome 15q25.1 in modifying lung cancer risk. Nat Commun 2018;9:3221. [Crossref] [PubMed]
- Grimm M, Cetindis M, Lehmann M, et al. Association of cancer metabolism-related proteins with oral carcinogenesis - indications for chemoprevention and metabolic sensitizing of oral squamous cell carcinoma? J Transl Med 2014;12:208. [Crossref] [PubMed]
- Huang WC, Jang TH, Tung SL, et al. A novel miR-365-3p/EHF/keratin 16 axis promotes oral squamous cell carcinoma metastasis, cancer stemness and drug resistance via enhancing beta5-integrin/c-met signaling pathway. J Exp Clin Cancer Res 2019;38:89. [Crossref] [PubMed]
- Sysel AM, Valli VE, Bauer JA. Immunohistochemical quantification of the cobalamin transport protein, cell surface receptor and Ki-67 in naturally occurring canine and feline malignant tumors and in adjacent normal tissues. Oncotarget 2015;6:2331-48. [Crossref] [PubMed]
- Chen SJ, Lian GD, Li JJ, et al. Tumor-driven like macrophages induced by conditioned media from pancreatic ductal adenocarcinoma promote tumor metastasis via secreting IL-8. Cancer Med 2018;7:5679-90. [Crossref] [PubMed]
- Muret K, Klopp C, Wucher V, et al. Long noncoding RNA repertoire in chicken liver and adipose tissue. Genet Sel Evol 2017;49:6. [Crossref] [PubMed]
- Kim W, Miguel-Rojas C, Wang J, et al. Developmental Dynamics of Long Noncoding RNA Expression during Sexual Fruiting Body Formation in Fusarium graminearum. MBio 2018; [Crossref] [PubMed]
- Cui C, Zhai D, Cai L, et al. Long Noncoding RNA HEIH Promotes Colorectal Cancer Tumorigenesis via Counteracting miR-939Mediated Transcriptional Repression of Bcl-xL. Cancer Res Treat 2018;50:992-1008. [Crossref] [PubMed]
- Işın M, Uysaler E, Ozgur E, et al. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front Genet 2015;6:168. [PubMed]
- Elkouris M, Kouroupi G, Vourvoukelis A, et al. Long Non-coding RNAs Associated With Neurodegeneration-Linked Genes Are Reduced in Parkinson’s Disease Patients. Front Cell Neurosci 2019;13:58. [Crossref] [PubMed]
- Liu T, Zhang H, Zheng J, et al. SPION-mediated miR-141 promotes the differentiation of HuAESCs into dopaminergic neuron-like cells via suppressing lncRNA-HOTAIR. J Cell Mol Med 2018;22:2299-310. [Crossref] [PubMed]
- Man H, Bi W. Expression of a Novel Long Noncoding RNA (lncRNA), GASL1, is Downregulated in Patients with Intracranial Aneurysms and Regulates the Proliferation of Vascular Smooth Muscle Cells In Vitro. Med Sci Monit 2019;25:1133-9. [Crossref] [PubMed]


