
LncRNA MEG3 inhibits the development of nasopharyngeal carcinoma by sponging miR-543 targeting KLF4
Introduction
Nasopharyngeal carcinoma (NPC) is a cancer of the epithelial cells of the pharynx (1). NPC epidemiology research showed a specific geographic distribution in southern China with a high incidence and prevalence rates (2). In addition, population-based analysis revealed a strong familial aggregation tendency among NPC patients, suggesting that genetic susceptibility can be critical in NPC tumorigenesis (3).
Cancer development is a complicated process of accumulated genetic and epigenetic changes over time that include activation of oncogenes or inhibition of tumor suppressor genes (4-6). The activation of oncogenes (activated from proto-oncogenes by chromosomal mutations) enhances abnormal cell biological behavior and results in cancerous cell changes and carcinogenesis (7). Tumor suppressor genes encode proteins and control cancer initiation and progression by cell division inhibition, apoptosis induction, damaged DNA repair and metastasis inhibition (8). However, the Human Genome Project (HPG) has revealed that only approximately 1.5% of the genome encodes protein (9). Most of the human genome can be transcribed but not translated, and since the first identification of regulatory non-coding RNAs (ncRNAs), molecular biology scientists have discovered that regulatory ncRNAs are involved in cell development and pathogenesis (10).
ncRNAs can be subcategorized into two classes by their functions: housekeeping (infrastructural) ncRNAs and regulatory ncRNAs (11). Housekeeping ncRNAs include ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA) and small nucleolar RNA (snoRNA), play pivotal roles in mediating vital cell functions (12). Regulatory ncRNAs can be divided into two subtypes based on their length: small ncRNAs (sncRNAs) with less than 200 nucleotides, which include miRNAs and snoRNAs, and long non-coding RNAs (lncRNAs), which are longer than 200 nucleotides (13,14). To date, most of the studies have investigated sncRNAs, especially miRNAs, as these molecules regulate target gene expression and function as oncogenes or tumor suppressor genes through their different targets (15). However, an increasing number of studies have shown evidence that lncRNA targets are related to the development and prognosis of different types of cancer, similar to miRNAs (16-21). Some lncRNAs, including LED, maternally expressed gene 3 (MEG3), GUARDIN and PTENP1, have been identified as tumor suppressors, and other lncRNAs, including MALAT1, HOTAIR, NORAD and PVT1, have been identified as tumor promoters (22).
LncRNAs have been shown to act as miRNA sponges to competitively bind miRNA sequences and suppress the interaction between miRNAs and mRNAs (23). For example, lncRNA MEG3 functions as a competitive endogenous RNA (ceRNA) of many different miRNAs, whereby downregulated MEG3 expression causes a change in miRNAs, subsequently affecting the expression levels of their target mRNAs (24). MEG3 can be found in various normal tissues, but gene expression is lost in multiplicate cancer cells and tissues (25,26). MEG3 is located at chromosome 14q32.3 in humans, and multiple mechanisms lead to alterations in MEG3 expression in tumors (27). Most of the studies have identified MEG3 as a tumor suppressor gene in many different cancers, such as glioma, gastric cancer, bladder cancer, ovarian cancer, cervical cancer, and lung cancer (28-33). MEG3 functions as a tumor suppressor by regulating tumor suppressor genes, inhibiting angiogenesis and miRNA control (34). Chak and colleagues found that MEG3 expression is decreased in NPC cells and plays a role in tumor suppression (35), but its detailed mechanism of tumor suppression in NPC cells is still unclear.
The bioinformatic software starBase 3.0 predicted that there are miRNA response elements (MRE) complementary to miR-543 in the MEG3 sequence, and Krüppel-like factor 4 (KLF4) was identified as a target gene of miR-543. There have been conflicting results regarding miR-543 expression levels in different types of cancers. MiR-543 is upregulated in colorectal cancer, gastric cancer, hepatocellular carcinoma, and prostate cancer, whereas it is decreased in ovarian cancer and endometrial cancer (36-42). Although miR-543 is expressed differently in cancers, it is clear that miR-543 is strongly involved in the development and progression of cancers. However, the role of miR-543 in nasopharyngeal cancer cells remains unknown.
In the present study, we determined MEG3 expression in nasopharyngeal cancer cells and its functional regulation in tumor development. In addition, we revealed the interactions among MEG3, miR-543 and KLF4 in nasopharyngeal cancer for the first time. This study evaluated MEG3 as a tumor suppressor and its underlying mechanisms in nasopharyngeal cancer.
Methods
Cell culture
Two NPC cell lines (C666-1 and TW03) and the human nasopharyngeal epithelial cell line NP69 were obtained from the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI-1640 medium (Invitrogen, USA) supplemented with 10% fetal bovine serum in a CO2 incubator (MCO-5AC, Sanyo, Japan) at 37 °C.
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from cells using NucleoZOL reagent (Machery-Nagel GmbH, Germany). The integrity of the RNA was assessed, and qRT-PCR was performed with an ABI 7500 system to detect the expression levels of MEG3, miR-543 and KLF4. The primer sequences were as follows: MEG3-F: 5'-GGGAAGGGACCTCGAATGTG-3', MEG3-R: 5'-CTGTCCCGTGGGAATAGGTG-3'; miR-543-F: 5'-CGAAACATTCGCGGTGCA-3'; miR-543-R: 5'-AGTGCAGGGTCCGAGGTATT-3', and miR-543-RT: 5'-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAGAAG-3'. GAPDH and U6 were used as internal controls, and the specific primers were as follows: GAPDH-F: 5'-GTCAAGGCTGAGAACGGGAA-3', GAPDH-R: 5'-AAATGAGCCCCAGCCTTCTC-3'; U6-F: 5'-CTCGCTTCGGCAGCACATATACT-3', U6-R: 5'-ACGCTTCACGAATTTGCGTGTC-3', and RT primer, 5'- AAAATATGGAACGCTTCACGAATTTG-3'. Relative gene expression was calculated using the 2–ΔΔCt method.
Cell transfection
si-MEG3, pcDNA-MEG3, miR-543 mimic, miR-543 inhibitor, si-KLF4, and pcDNA-KLF4 were obtained from Genepharma Company (Shanghai, China). Transfection was performed using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s protocols.
Cell proliferation assay
Cell proliferation was measured with the cell counting kit-8 (CCK-8) following the manufacturer’s instructions. After 48 hours of transfection, C666-1 cells were seeded into a 96-well plate. Then, 100 µL of 10% CCK-8 solution (CK04, Dojindo) was added into each well at 0, 24, 48, 72 and 96 h after transfection and incubated for 2 h at 37 °C. The absorbance at OD450 nm was measured by a microplate reader.
Cell apoptosis assay
Cell apoptosis was assessed by flow cytometry analysis. Cells were cultured overnight and transfected when they reached 80–90% confluence. Cells were cultured in a 37 °C incubator supplemented with 5% CO2 and digested with trypsin without EDTA. The cells were harvested after centrifugation and washed with PBS. The collected cells were mixed with binding buffer and then incubated with Annexin V-FITC and PI staining solutions for approximately 15 min in the dark at room temperature. The apoptosis rate was counted using flow cytometry.
Cell migration and invasion assays
Each group of transfected cells (1×105 cells/mL) were diluted in RPMI-1640 medium without serum and seeded into the upper chamber. The lower chamber was filled with RPMI-1640 medium containing 20% FBS. After incubation at 37 °C for 24 h, the cells in the upper chambers were removed. Cells were fixed and stained with crystal violet. Then, randomly selected fields were imaged, and migrated or invaded cells were counted.
Dual luciferase assay
The target genes of miR-543 were predicted using a miRNA database (TargetScan: http://www.targetscan.org/). The putative binding sequences of MEG3-WT (5'GAGUAAUG-GUAGUGAAUGUUU3') and MEG3-MUT (5'GAGUAAUG-GUAGUCAGAUCAU3'), were cloned into the pcDNA3.1 vector and then cotransfected with miR-543 mimic or negative control in C666-1 cells using Lipofectamine 2000 (Invitrogen, USA). After 36 h of transfection, firefly and Renilla signals were analyzed with a Dual-Luciferase Reporter Assay System (Promega, USA). Luciferase activity was quantified by a microplate reader.
Western blotting
Total protein was isolated using RIPA buffer (Beyotime, Shanghai, China), separated by 10% SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane. Quantification of protein was analyzed using the Bradford method. Visualization of target protein bands on the PVDF membranes were performed with a Pierce ECL kit. The following antibodies were used and were diluted as follows: anti-KLF (1:1,000, Proteintech, USA), anti-Bcl-2 (1:1,000, Cell Signaling Technology, USA) and anti-Bax (1:1,000, Cell Signaling Technology). Anti-β-actin (1:1,000; Cell Signaling Technology) was used as an internal control.
Statistical analysis
The data are expressed as the mean ± standard deviation (SD) and were analyzed by the statistical software SPSS 16.0 (SPSS, Inc., Chicago, IL, USA). Statistical significance was determined using analysis of variance (ANOVA). P value <0.05 was considered to indicate a statistically significant difference, and P value <0.001 was considered to indicate a statistically highly significant difference.
Results
MEG3 was decreased and miR-543 was increased in human NPC cell lines
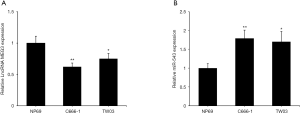
MEG3 and miR-543 expression levels in NPC cell lines (C666-1 and TW03) and in the normal nasopharyngeal epithelial cell line NP69 were determined by qRT-PCR. Compared with those in the normal control, the MEG3 levels in NPC cell lines were decreased (Figure 1A), whereas miR-543 levels were increased (Figure 1B). Ultimately, the C666-1 cell line was selected for the following investigations.
MEG3 affected NPC cell biological behavior
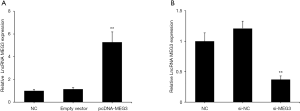
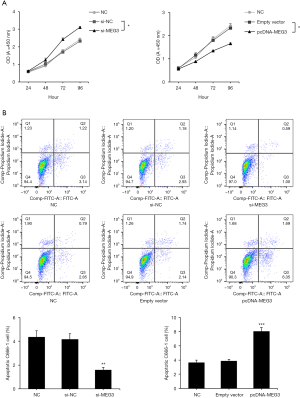
To explore the effects of MEG3 on NPC cell biological behavior, pcDNA-MEG3 or si-MEG3 was transfected into C666-1 cells. qRT-PCR results showed that MEG3 expression was markedly increased by pcDNA-MEG3 (Figure 2A, P<0.01) and reduced by si-MEG3 (Figure 2B, P<0.01). Cell proliferation was assessed by the CCK-8 assay. The si-MEG3 group showed gradually increased OD values compared to the control, whereas the OD values in the pcDNA-MEG3 group decreased in a time-dependent manner (Figure 3A). In addition, apoptotic cells were measured by flow cytometry assay, and the results revealed that pcDNA-MEG3 increased the number of cells in the early and late stages of apoptosis, whereas si-MEG3 decreased apoptosis and the number of dead cells (Figure 3B). These findings confirmed that si-MEG3 enhanced proliferation and suppressed apoptosis in C666-1 cells, whereas pcDNA-MEG3 decreased cell proliferation and enhanced cell apoptosis.
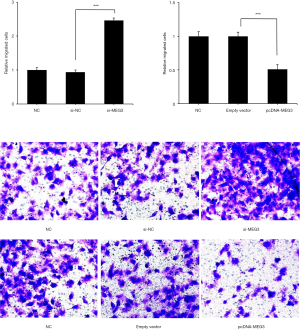
Migration and invasion assays were performed in Transwells. Si-MEG3 significantly increased cell migration, whereas pcDNA-MEG3 significantly decreased cell migration (Figure 4, P<0.001). Similarly, invaded cells were significantly increased by si-MEG3 and significantly decreased by pcDNA-MEG3 (Figure 5, P<0.001). These results indicated that pcDNA-MEG3 decreased cell migration and invasion, whereas si-MEG3 increased cell migration and invasion.

MEG3 directly sponged miR-543 in C666-1 cells
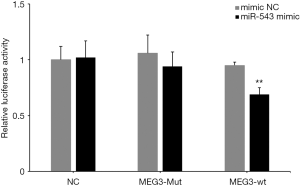
The preliminary investigation predicted a correlation between miRNA-543 and MEG3. Figure 1A,B shows a significant upregulation of miR-543 expression and significant downregulation of MEG3 in NPC cell lines. Bioinformatics prediction revealed that MEG targeted miR-543. A luciferase assay was used to assess the relationship between miRNA-543 and MEG3. Thus, MEG3 wild-type and mutant luciferase reporter vectors were constructed and transfected into cells. The luciferase activity results indicated that the miR-543 mimic markedly decreased the luciferase activity in the MEG3 wild-type reporter group compared with the NC group. In contrast, the miR-543 mimic had no detectable effect on the luciferase activity in the MEG3 mutant group (Figure 6). These data suggest that MEG3 is a miR-543 target in NPC cells and that there is negative regulation between miR-543 and MEG3 in NPC cells.
MEG3 increased KLF4 expression by sponging miR-543
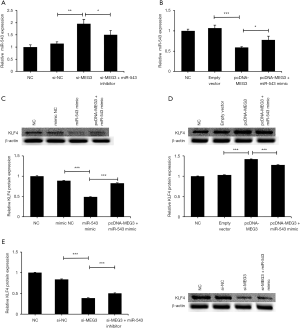
Some known lncRNAs act as ceRNAs (also called ‘miRNA sponges’) by directly binding to miRNAs to inhibit miRNA expression (43). Downregulation of MEG3 greatly elevated miR-543 levels, but the cotransfection of miR-543 inhibitor with si-MEG3 markedly inhibited the promotion effect of si-MEG3 on miR-543 expression (Figure 7A). The overexpression of MEG3 dramatically decreased the miR-543 expression level, but cotransfection of pcDNA-MEG3 and miR-543 mimic reversed the inhibitory effect of pcDNA-MEG3 on the expression of miR-543 (Figure 7B). Upregulation of miR-543 inhibited KLF4 expression in C666-1 cells, whereas the cotransfection of miR-543 mimic with pcDNA-MEG3 attenuated the suppressor function of miR-543 on KLF4 expression in C666-1 cells (Figure 7C). The overexpression of MEG3 markedly promoted the KLF4 protein level, whereas the cotransfection of miR-543 mimic with pcDNA-MEG3 reversed the promotion effect of pcDNA-MEG3 on KLF4 expression (Figure 7D). Downregulation of MEG3 significantly decreased KLF4 protein levels, whereas cotransfection of miR-543 inhibitor with si-MEG3 markedly relieved the suppressive effect of si-MEG3 on KLF4 expression (Figure 7E).
MEG3 acted as a tumor suppressor by regulating KLF4 expression in NPC
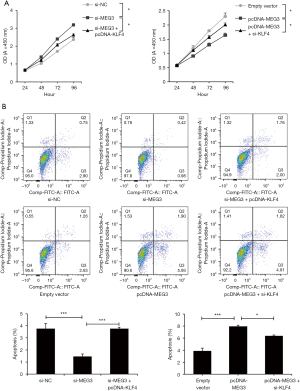
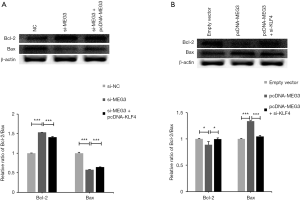
To further study whether KLF4 mediated the function of MEG3 in the progression of NPC, functional rescue experiments were performed. We found that si-MEG3 promoted cell proliferation and that pcDNA-MEG3 reduced cell proliferation; however, the cotransfection of pcDNA-KLF4 reversed the promoting effect of si-MEG3 on C666-1 cell proliferation, and the cotransfection of si-KLF4 relieved the inhibitory effect of pcDNA-MEG3 on cell proliferation (Figure 8A). Si-MEG3 suppressed cell apoptosis, whereas si-MEG3 + pcDNA-KLF4 alleviated the inhibitory function of si-MEG3 in C666-1 cell apoptosis (Figure 8B). Conversely, we found that pcDNA-MEG3 induced cell apoptosis, whereas pcDNA-MEG3 and si-KLF4 cotransfection inhibited the promoting effect of pcDNA-MEG3 on cell apoptosis (Figure 8B). In addition, the apoptosis-related Bcl-2 and Bax proteins were analyzed by western blotting. The Bcl-2 family includes Bcl-2, which induces apoptosis, and Bax, which promotes apoptosis, is known to inhibit or enhance apoptosis and has become a novel cancer therapeutic to control apoptosis (44-46). In particular, high Bax mRNA expression can be used as a good prognostic indicator of survival rate in NPC patients (47). Knockdown of MEG3 increased Bcl-2 expression and reduced Bax expression; however, forced expression of KLF4 reversed these effects in C666-1 cells (Figure 9A). Overexpression of MEG3 decreased Bcl-2 expression and elevated Bax expression; however, reduced expression of KLF4 reversed these effects in C666-1 cells (Figure 9B). Consequently, the role of MEG3 in C666-1 cell proliferation and apoptosis was regulated, at least in part, by KLF4.
Discussion
NPC has a high prevalence and mortality rate in Southeast Asia, and the main reason for the high mortality is the late detection because of the lack of specific symptoms (48). Patients at an advanced stage of NPC have poor prognosis and a high recurrence rate, even with aggressive treatment (49). Thus, recent studies have been conducted to elucidate the molecular basis of NPC carcinogenesis, discover reliable biomarkers of NPC and develop effective molecularly targeted therapies.
After the genome project showed that only approximately 1.5% of the genome encoded protein, Encyclopedia of DNA Elements (ENCODE) project researchers revealed that 80.4% of the human genome is engaged in biochemical events such as binding to protein or chromatin structures, and discovered that regulatory regions can act differently according to the cell type (50). LncRNAs are thought to be involved in various disease etiologies and regulate target gene expression (51). A growing number of studies have shown that lncRNAs can be oncogenes or tumor suppressors in carcinogenesis (52). The first identification of MEG3 involvement in carcinogenesis suppression was conducted by Zhang et al. in 2003 (53), and subsequent investigations identified MEG3 as a tumor suppressor in multiplicate cancer cells. MEG3 physiological and pathological processes in cancer cells have been under continuous investigation, but the role of MEG3 in NPC progression remains unknown.
This research aimed to analyze the MEG3, miR-543 and KLF4 expression and interactions in NPC cells and the effects on NPC cell progression and carcinogenesis. MiR-543 can have oncogenic or tumor suppressive effects depending on the cancer cell type. KLF4 is a known target gene of miR-543 that is involved in cell proliferation and functions as a tumor suppressor and prognostic indicator in cancer (36,54,55). In the present study, the MEG3 expression level was decreased in NPC cells, and pleiotropic regulation of NPC cell biological behavior was observed. MEG3 overexpression decreased cell proliferation, migration and invasion but promoted cell apoptosis. MEG3 exerted a tumor suppressive role by targeting miR-543 and acting as a ceRNA. Furthermore, KLF4 was confirmed as a targeted gene of miR-543 in NPC cells, and this was consistent with previous studies. KLF4 was downregulated by miR-543, and KLF4 expression inhibited C666-1 cell proliferation and induced cell apoptosis. KLF4 in NPC cells regulated Bcl-2 and Bax protein expression, indicating that KLF4 may be a prognostic factor for NPC.
In conclusion, our findings suggest a tumor suppressive role of MEG3 through the MEG3-miR-543-KLF4 axis in NPC cells. Decreased MEG3 was negatively correlated with miR-543 expression and could reverse the decrease in KLF4 expression caused by miR-543 overexpression, which was associated with NPC progression. MEG3 may represent a new therapeutic biomarker for NPC targeted treatment.
Acknowledgments
Funding: This work was supported by grants from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.41). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. An ethics statement was no required for this research as we did the experiment only using cells from the Chinese Academy of Sciences (Shanghai, China)
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Brennan B. Nasopharyngeal carcinoma. Orphanet J Rare Dis 2006;1:23. [Crossref] [PubMed]
- Cao SM, Simons MJ, Qian CN. The prevalence and prevention of nasopharyngeal carcinoma in China. Chin J Cancer 2011;30:114-9. [Crossref] [PubMed]
- Huang SF, Hsiao JH, Young CK, et al. Familial aggregation of nasopharyngeal carcinoma in Taiwan. Oral Oncol 2017;73:10-5. [Crossref] [PubMed]
- Cooper GM, Hausman RE. The cell: a molecular approach. 7th ed. Washington DC: ASM Press, 2000.
- Barrett JC, Wiseman RW. Cellular and molecular mechanisms of multistep carcinogenesis: relevance to carcinogen risk assessment. Environ Health Perspect 1987;76:65-70. [Crossref] [PubMed]
- Bommer UA, Vine KL. Cancer biology: molecular and genetic basis. 2014. Available online: http://ro.uow.edu.au/cgi/viewcontent.cgi?article=1495&context=ihmri
- Botezatu A, Iancu IV, Popa O, et al. Mechanisms of oncogene activation. In: Bulgin D. editor. New aspects in molecular and cellular mechanisms of human carcinogenesis. Croatia: IntechOpen, 2016.
- Sun W, Yang J. Functional mechanisms for human tumor suppressors. J Cancer 2010;1:136-40. [Crossref] [PubMed]
- Lander ES. Initial impact of the sequencing of the human genome. Nature 2011;470:187-97. [Crossref] [PubMed]
- Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol 2017;1008:1-46. [Crossref] [PubMed]
- Kim ED, Sung S. Long noncoding RNA: unveiling hidden layer of gene regulatory networks. Trends Plant Sci 2012;17:16-21. [Crossref] [PubMed]
- Hansji H, Leung EY, Baguley BC, et al. Keeping abreast with long non-coding RNAs in mammary gland development and breast cancer. Front Genet 2014;5:379. [Crossref] [PubMed]
- Li Z, Shen J, Chan MT, et al. TUG 1: a pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif 2016;49:471-5. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- Wright MW, Bruford EA. Naming 'junk': human non-protein coding RNA (ncRNA) gene nomenclature. Hum Genomics 2011;5:90-8. [Crossref] [PubMed]
- Chakravarty D, Sboner A, Nair SS, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun 2014;5:5383. [Crossref] [PubMed]
- Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 2014;5:2318-29. [Crossref] [PubMed]
- Augoff K, McCue B, Plow EF, et al. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer 2012;11:5. [Crossref] [PubMed]
- Zheng HT, Shi DB, Wang YW, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol 2014;7:3174-81. [PubMed]
- Cao S, Liu W, Li F, et al. Decreased expression of lncRNA GAS5 predicts a poor prognosis in cervical cancer. Int J Clin Exp Pathol 2014;7:6776-83. [PubMed]
- Yang YR, Zang SZ, Zhong CL, et al. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol 2014;7:6929-35. [PubMed]
- Sanchez Calle A, Kawamura Y, Yamamoto Y, et al. Emerging roles of long non-coding RNA in cancer. Cancer Sci 2018;109:2093-100. [Crossref] [PubMed]
- Fernandes JCR, Acuña SM, Aoki JI, et al. Long non-coding RNAs in the regulation of gene expression: physiology and disease. Noncoding RNA 2019; [Crossref] [PubMed]
- Moradi MT, Fallahi H, Rahimi Z. Interaction of long noncoding RNA MEG3 with miRNAs: a reciprocal regulation. J Cell Biochem 2019;120:3339-52. [Crossref] [PubMed]
- He Y, Luo Y, Liang B, et al. Potential applications of MEG3 in cancer diagnosis and prognosis. Oncotarget 2017;8:73282-95. [PubMed]
- Wang Q, Li M, Shen Z, et al. The long non-coding RNA MEG3/miR-let-7c-5p axis regulates ethanol-induced hepatic steatosis and apoptosis by targeting NLRC5. Front Pharmacol 2018;9:302. [Crossref] [PubMed]
- Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol 2012;48:R45-53. [Crossref] [PubMed]
- Sun M, Xia R, Jin F, et al. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol 2014;35:1065-73. [Crossref] [PubMed]
- Qin R, Chen Z, Ding Y, et al. Long non-coding RNA MEG3 inhibits the proliferation of cervical carcinoma cells through the induction of cell cycle arrest and apoptosis. Neoplasma 2013;60:486-92. [Crossref] [PubMed]
- Ying L, Huang Y, Chen H, et al. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst 2013;9:407-11. [Crossref] [PubMed]
- Su L, Han D, Wu J, et al. Skp2 regulates non-small cell lung cancer cell growth by Meg3 and miR-3163. Tumour Biol 2016;37:3925-31. [Crossref] [PubMed]
- Lu KH, Li W, Liu XH, et al. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 2013;13:461. [Crossref] [PubMed]
- Xiu YL, Sun KX, Chen X, et al. Upregulation of the lncRNA Meg3 induces autophagy to inhibit tumorigenesis and progression of epithelial ovarian carcinoma by regulating activity of ATG3. Oncotarget 2017;8:31714-25. [Crossref] [PubMed]
- Al-Rugeebah A, Alanazi M, Parine NR. MEG3: an oncogenic long non-coding RNA in different cancers. Pathol Oncol Res 2019;25:859-74. [Crossref] [PubMed]
- Chak WP, Lung RW, Tong JH, et al. Downregulation of long non-coding RNA MEG3 in nasopharyngeal carcinoma. Mol Carcinog 2017;56:1041-54. [Crossref] [PubMed]
- Zhai F, Cao C, Zhang L, et al. miR-543 promotes colorectal cancer proliferation and metastasis by targeting KLF4. Oncotarget 2017;8:59246-56. [Crossref] [PubMed]
- Li J, Dong G, Wang B, et al. miR-543 promotes gastric cancer cell proliferation by targeting SIRT1. Biochem Biophys Res Commun 2016;469:15-21. [Crossref] [PubMed]
- Song N, Liu H, Ma X, et al. Placental growth factor promotes metastases of ovarian cancer through MiR-543-regulated MMP7. Cell Physiol Biochem 2015;37:1104-12. [Crossref] [PubMed]
- Yu L, Zhou L, Cheng Y, et al. MicroRNA-543 acts as an oncogene by targeting PAQR3 in hepatocellular carcinoma. Am J Cancer Res 2014;4:897-906. [PubMed]
- Bing L, Hong C, Li-Xin S, et al. MicroRNA-543 suppresses endometrial cancer oncogenicity via targeting FAK and TWIST1 expression. Arch Gynecol Obstet 2014;290:533-41. [Crossref] [PubMed]
- Du Y, Liu XH, Zhu HC, et al. MiR-543 promotes proliferation and epithelial-mesenchymal transition in prostate cancer via targeting RKIP. Cell Physiol Biochem 2017;41:1135-46. [Crossref] [PubMed]
- Fan C, Lin Y, Mao Y, et al. MicroRNA-543 suppresses colorectal cancer growth and metastasis by targeting KRAS, MTA1 and HMGA2. Oncotarget 2016;7:21825-39. [PubMed]
- Huang Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med 2018;22:5768-75. [Crossref] [PubMed]
- Hata AN, Engelman JA, Faber AC. The BCL2 family: key mediators of the apoptotic response to targeted anticancer therapeutics. Cancer Discov 2015;5:475-87. [Crossref] [PubMed]
- Skommer J, Brittain T, Raychaudhuri S. Bcl-2 inhibits apoptosis by increasing the time-to-death and intrinsic cell-to-cell variations in the mitochondrial pathway of cell death. Apoptosis 2010;15:1223-33. [Crossref] [PubMed]
- Wang Q, Zhang L, Yuan X, et al. The relationship between the Bcl-2/Bax proteins and the mitochondria-mediated apoptosis pathway in the differentiation of adipose-derived stromal cells into neurons. PLoS One 2016;11:e0163327. [Crossref] [PubMed]
- Kontos CK, Fendri A, Khabir A, et al. Quantitative expression analysis and prognostic significance of the BCL2-associated X gene in nasopharyngeal carcinoma: a retrospective cohort study. BMC Cancer 2013;13:293. [Crossref] [PubMed]
- Abdullah B, Alias A, Hassan S. Challenges in the management of nasopharyngeal carcinoma: a review. Malays J Med Sci 2009;16:50-4. [PubMed]
- Cheng SH, Tsai SY, Yen KL, et al. Concomitant radiotherapy and chemotherapy for early-stage nasopharyngeal carcinoma. J Clin Oncol 2000;18:2040-5. [Crossref] [PubMed]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57-74. [Crossref] [PubMed]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009;10:155-9. [Crossref] [PubMed]
- Zeng C, Xu Y, Xu L, et al. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer 2014;14:693. [Crossref] [PubMed]
- Zhang X, Zhou Y, Mehta KR, et al. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab 2003;88:5119-26. [Crossref] [PubMed]
- Garrett-Sinha LA, Eberspaecher H, Seldin MF, et al. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem 1996;271:31384-90. [Crossref] [PubMed]
- Hu W, Jia Y, Xiao X, et al. KLF4 downregulates hTERT expression and telomerase activity to inhibit lung carcinoma growth. Oncotarget 2016;7:52870-87. [PubMed]


