
Copy number variations of MMP-9 are prognostic biomarkers for hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the most common primary malignancy of the liver. The 5-year survival rate of HCC patients after curative resection was 5–9% from the time of clinical diagnosis (1). In 2012, around 782,500 new patients were reported with liver cancer and 745,500 deaths occurred in the world, and China accounted for about 50% of them (2). With the recent progress in modern medical technologies and the early diagnosis, the resection rate of HCC has been significantly improved. However, a high probability of recurrence after resection has remained a significant challenge in HCC therapy (3). The poor prognosis of HCC is related to high invasion and metastatic capacities of cancer cells (4), which are closely predicted by alpha-fetoprotein (AFP), tumor size, differentiation, clinical stage, and microvascular invasion (MVI) (3).
Matrix metalloproteinase-9 (MMP-9) is a matrixin, a class of enzymes that belong to the zinc-metalloproteinases family. A previous study reported that MMPs played a vital role in cancer invasion and during different stages of cancer progression (5). MMP-9 is highly expressed in HCC, and it participates in angiogenesis by degrading the environmental extracellular matrix and basement membrane (6). Another research showed that MMP-9 was expressed at a higher level in HCC tissues than in adjacent normal tissues, and MMP-9 expression was significantly higher in patients with distant metastases or portal vein invasion, indicating that MMP-9 played a crucial role in the invasion and the metastasis of HCC (7). Several studies were undertaken on MMP-9 polymorphism of cardio-cerebrovascular disease, and the achieved results confirmed an essential role of MMP-9 in myocardial infarction (8), temporomandibular disorders (9), intracranial hemorrhage in patients with brain arteriovenous malformation (10), and thoracic aortic dissection (11). However, the role of MMP-9 polymorphism in HCC is still unknown.
Our genome contains many intermediate size copy number changes, gains, and losses, called copy number variations (CNVs) (12). CNVs can reshape gene structure, modulate gene expression, and contribute to significant phenotypic variation (13). These genomic alterations can range from small insertions or deletions (less than 10 kb) to large ones (over 1 Mb) (14). CNVs are one of the most common genetic variations in the human genome as well as being an important molecular mechanism of pathogenesis in different human diseases such as cancer (15). Digital polymerase chain reaction (dPCR) offers a quantitative method to measure the abundance of a target molecule without requiring a calibration curve, leading to accurate copy number results (16).
In the present study, tumor tissues and adjacent normal tissues were collected from HCC patients in Ningbo Medical Center Lihuili Hospital. This paper aimed to investigate the contribution of MMP-9 CNVs to HCC prognosis and recurrence.
Methods
Patient and public involvement
The patients were gathered in Ningbo Medical Center Lihuili Hospital between January 2016 and December 2018. All patients were diagnosed by ultrasonography and computed tomography (CT), and histologically diagnosed with HCC. Subjects who had congenital heart disease or other cancers were excluded from the study. The results would be disseminated to each of the participants through the patient’s forum.
Origin of specimens
The HCC tumor tissues (n=35), as well as adjacent non-tumor tissues (the tissues from the edge of tumor tissues larger than 2 cm, n=35), were obtained from patients who underwent surgical resections. The HCC tumor tissues and adjacent non-tumor tissues were both confirmed by pathologic diagnoses. The clinical characteristics including the hepatitis B virus, alpha-fetoprotein (AFP), tumor size, differentiation, microvascular invasion (MVI), and clinical stage are collected for the patients. This study was approved by the Ethics Committee of Ningbo Medical Center Lihuili Hospital (Project Identification Code: DYLL2018028), and informed consent was obtained from all patients before the study.
Genomic DNA isolation and TaqMan® copy number assay
Genomic DNA (gDNA) was isolated from tumor tissues and adjacent-normal tissues using the QIAamp DNA Mini Kit (QIAGEN, Germany). The concentration of the purified gDNA was determined by Infinite M200 PRO (TECAN, Switzerland). The FAM™ dye-labeled TaqMan® Copy Number Reference Assay for MMP-9 (Cat. No. 4400291) was duplexed with the VIC® dye-labeled TaqMan® Copy Number Reference Assay for RNase P (RPPH1, Cat. No. 4403326).
MMP-9 copy number assay
The copy number of MMP-9 was measured in each gDNA sample using the QuantStudio™ 3D Digital PCR System (Life Technologies Corporation, NY, USA). RPPH1 and MMP-9 signals were amplified in each PCR. A total volume of 16 µL of PCR was prepared for each sample, containing QuantStudio® 3D Digital PCR Master Mix, TaqMan® Copy Number Assay for MMP-9, RNase P Reference Assay, and gDNA sample. The PCR was then loaded into the QuantStudioTM 3D Digital PCR 20K chip, which was loaded onto the Dual Flat Block GeneAmp PCR System 9700. The PCR program was as follows: initial melting at 96 °C for 10 minutes followed by 39 cycles at 60 °C for 2 minutes, 98 °C for 30 seconds, and 2 holds at 60 °C for 2 minutes. After PCR amplification, chips were read on the QuantStudio® 3D Digital PCR Instrument. Absolute quantification data were exported from QuantStudio™ 3D AnalysisSuite™ Software (Life Technologies Corporation, NY, USA). MMP-9 dPCR was duplexed with RPPH1 as the baseline control (17). The copy number of MMP-9 was calculated with the following equation: MMP-9 copy number = raw MMP-9 number/(raw RPPH1 number/2). As the number of copies of MMP-9 was 2 in the normal population, the MMP-9 CNV (ΔCN value) in HCC patients was calculated by the following equation: ΔCN value =|measured MMP-9 copies – 2|.
RNA isolation and quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Total RNAs were extracted from tumor tissues and adjacent non-tumor tissues using TRIzol reagent (Invitrogen, CA, USA). RNA samples were measured by optical density at 260 nm and reversely transcribed using a PrimeScript RT reagent Kit (Takara, Japan). Quantitative PCR (Q-PCR) was performed in the ViiA 7 Q-PCR System (Applied Biosystems Inc., CA, USA) using SYBR® Green PCR Kit (QIAGEN, Germany). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a controller. The primer sequences were 5'-GCCTGCCACTTCCCCTTCAT-3' for forward primer of MMP-9, 5'-CAGAAGCCAAACCGGTCGTC-3' for reverse primer of MMP-9, 5'-GGGAAATCGTGCGTGACAT-3' for forward primer of GAPDH, 5'-TGTTGCTGTAGCCAAATTCGTT-3' for reverse primer of GAPDH. The PCR program was as follows: initial denaturation at 95 °C for 2 minutes followed by 45 cycles at 94 °C for 10 seconds, 60 °C for 10 seconds, and 72 °C for 40 seconds. The fold variations in mRNAs were normalized to GAPDH and calculated using the 2-ΔΔCt method.
Statistical analysis
The statistical analysis was performed using GraphPad Prism software for Windows, version 5.01 (GraphPad Software Inc., CA, USA). The differences between groups were analyzed by two-tailed t-test. Quantitative data were compared using a one-way analysis of variance (ANOVA) or the Kruskal Wallis test. Spearman’s rank correlation coefficient (r) was used to determine a relationship between MMP9 copy number and differentiation in HCC. Data were analyzed using the Mann-Whitney U test. Receiver operating characteristic (ROC) curves were established to evaluate the diagnostic value for the disease. A two-sided P<0.05 was defined as statistically significant.
Results
Characteristics of the tested participants
The clinical phenotypes of tumors and treatments for HCC patients are listed in Table 1. There were 12 patients in clinical stage I, 12 patients in the clinical stage II, 5 patients in clinical stage IIIa, 5 patients in the clinical stage IIIb, 1 patient in clinical stage IIIc. All the patients were treated by surgical resection. As shown in Table 2, there were 29 patients with positive hepatitis B virus and 20 patients with positive AFP. Among the participants, 2 patients were well differentiated, 21 patients were moderately differentiated, and 12 patients were poorly differentiated. The MVI test showed that 10 patients were not MVI, 16 patients were M1 (low risk), 9 patients were M2 (high risk). The histopathological pictures of tumor tissues and adjacent non-tumor tissues are shown in Figure 1.
Table 1
| Patients | Age | Gender | Grading of tumor | Treatment |
|---|---|---|---|---|
| Patient 1 | 43 | Male | IIIa | Surgical resection |
| Patient 2 | 56 | Male | II | Surgical resection |
| Patient 3 | 48 | Male | II | Surgical resection |
| Patient 4 | 45 | Male | I | Surgical resection |
| Patient 5 | 82 | Male | II | Surgical resection |
| Patient 6 | 61 | Male | I | Surgical resection |
| Patient 7 | 54 | Male | II | Surgical resection |
| Patient 8 | 65 | Male | IIIb | Surgical resection |
| Patient 9 | 60 | Male | I | Surgical resection |
| Patient 10 | 67 | Male | I | Surgical resection |
| Patient 11 | 66 | Male | I | Surgical resection |
| Patient 12 | 69 | Male | I | Surgical resection |
| Patient 13 | 60 | Male | I | Surgical resection |
| Patient 14 | 42 | Male | IIIb | Surgical resection |
| Patient 15 | 76 | Female | II | Surgical resection |
| Patient 16 | 69 | Female | I | Surgical resection |
| Patient 17 | 69 | Male | I | Surgical resection |
| Patient 18 | 51 | Male | IIIc | Surgical resection |
| Patient 19 | 61 | Male | II | Surgical resection |
| Patient 20 | 43 | Male | IIIa | Surgical resection |
| Patient 21 | 65 | Male | IIIb | Surgical resection |
| Patient 22 | 43 | Male | IIIb | Surgical resection |
| Patient 23 | 58 | Male | IIIb | Surgical resection |
| Patient 24 | 71 | Female | II | Surgical resection |
| Patient 25 | 57 | Male | I | Surgical resection |
| Patient 26 | 58 | Male | II | Surgical resection |
| Patient 27 | 64 | Male | IIIa | Surgical resection |
| Patient 28 | 47 | Male | I | Surgical resection |
| Patient 29 | 71 | Male | IIIa | Surgical resection |
| Patient 30 | 64 | Male | IIIa | Surgical resection |
| Patient 31 | 64 | Male | II | Surgical resection |
| Patient 32 | 63 | Male | II | Surgical resection |
| Patient 33 | 69 | Male | II | Surgical resection |
| Patient 34 | 55 | Male | I | Surgical resection |
| Patient 35 | 68 | Male | II | Surgical resection |
Table 2
| Characteristics | No. of cases |
|---|---|
| Age (years) | 60.11±10.10 |
| Gender | |
| Male | 32 |
| Female | 3 |
| Hepatitis B virus | |
| Positive | 29 |
| Negative | 6 |
| AFP | |
| Positive | 20 |
| Negative | 15 |
| Tumor size, cm | |
| ≥5 | 15 |
| <5 | 20 |
| Differentiation | |
| Well | 2 |
| Moderate | 21 |
| Poor | 12 |
| MVI | |
| M0 | 10 |
| M1 | 16 |
| M2 | 9 |
| Clinical stage | |
| I | 12 |
| II | 12 |
| IIIa | 5 |
| IIIb | 5 |
| IIIc | 1 |
AFP, alpha fetoprotein; MVI, microvascular invasion. M0, no MVI; M1 (low-risk), MVI of <5 and at ≤1 cm away from the adjacent liver tissues; M2 (high-risk), MVI of >5 or at >1 cm away from the adjacent liver tissues.
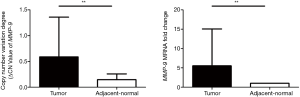
MMP-9 CNVs degree (ΔCN value) and mRNA fold change in HCC tumor tissues and adjacent non-tumor tissues
We measured the copy number of MMP-9 in gDNAs isolated from tumor tissues (n = 35) as well as adjacent non-tumor tissues (n=35) in HCC patients using dPCR. Our results showed that MMP-9 CNVs (ΔCN value) were significantly higher in tumor tissues (0.589±0.770) than in adjacent normal tissues (0.146±0.112, P=0.002, Figure 2). We also found that the fold change of MMP-9 mRNA was significantly higher in tumor tissues (5.521±9.545) than adjacent non-tumor tissues (1.000±0.000, P=0.0047).
Relationship between MMP-9 CNVs and clinicopathological factors in HCC patients
Our results showed that there were 15 HCC patients with CNVs in tumor tissues (CNV group) and 20 HCC patients without CNVs in tumor tissues (non-CNV group). Therefore, we compared the clinicopathological factors between CNVs group and non-CNVs group. As shown in Table 3, significant differences of tissue AFP expression (P=0.015), tumor size (P<0.001), differentiation (P<0.001), MVI (P=0.009), and clinical stage (P<0.001) were found between CNV group and non-CNV group. In contrast, there was no significant differences between CNV group and non-CNV group for the other clinicopathological factors, including age (P=0.659), gender (P=0.446), hepatitis B virus (P=0.351) and MMP-9 mRNA expression level (P=0.430).
Table 3
| Characteristics | CNVs group, n=15 | Non-CNVs group, n=20 | P value |
|---|---|---|---|
| Age (mean ± SD) | 61±8.960 | 59.45±11.057 | 0.659 |
| Gender (n, %) | 0.446 | ||
| Male | 14 (93.3) | 18 (90.0) | |
| Female | 1 (6.7) | 2 (10.0) | |
| Hepatitis B virus (n, %) | 0.351 | ||
| Positive | 12 (80.0) | 17 (85.0) | |
| Negative | 3 (20.0) | 3 (15.0) | |
| AFP (n, %) | 0.015 | ||
| Positive | 10 (66.7) | 10 (50.0) | |
| Negative | 5 (33.3) | 10 (50.0) | |
| Tumor size, cm (n, %) | <0.001 | ||
| ≥5 | 9 (60.0) | 6 (30.0) | |
| <5 | 6 (40.0) | 14 (70.0) | |
| Differentiation (n, %) | <0.001 | ||
| Well | 0 (0) | 2 (10.0) | |
| Moderate | 8 (53.3) | 13 (65.0) | |
| Poor | 7 (46.7) | 5 (25.0) | |
| MVI (n, %) | 0.009 | ||
| M0 | 2 (13.3) | 8 (40.0) | |
| M1 | 8 (53.3) | 8 (40.0) | |
| M2 | 5 (33.3) | 4 (20.0) | |
| Clinical stage (n, %) | <0.001 | ||
| I | 4 (26.7) | 8 (40.0) | |
| II | 5 (33.3) | 7 (35.0) | |
| IIIa | 2 (13.3) | 3 (15.0) | |
| IIIb | 3 (20.0) | 2 (10.0) | |
| IIIc | 1 (6.7) | 0 (0) | |
| MMP-9 mRNA expression | 4.76±2.494 | 4.13±2.18 | 0.430 |
AFP, alpha fetoprotein; MVI, microvascular invasion. M0, no MVI; M1 (low-risk), MVI of <5 and at ≤1 cm away from the adjacent liver tissues; M2 (high-risk), MVI of >5 or at >1 cm away from the adjacent liver tissues. *, The P value is calculated according to the percentage. Data were analyzed using one-way analysis of variance (ANOVA), the Kruskal Wallis test or Mann-Whitney U test. The cut-off concentrations used to distinguish positive and negative results were 20 μg/L for AFP and 0.9 S/CO for hepatitis B virus.
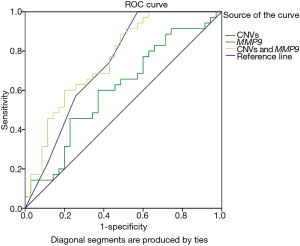
Additionally, our ROC analysis for prediction potential of MMP-9 CNVs and expression levels for HCC showed that MMP-9 CNVs and MMP-9 expression were significantly associated with HCC [CNVs: P=0.001, area under curve (AUC) =0.74; MMP-9 expression: P=0.047, AUC =0.64, Figure 3). Moreover, the combined ROC showed a more enhanced diagnostic ability of MMP-9 CNVs and expression for HCC (P<0.0001, AUC =0.76, Figure 3).
Discussion
Our study indicated that the copy number and transcription level of MMP-9 was significantly higher in tumor tissues than in adjacent normal tissues. Also, MMP-9 CNVs were shown to be associated with several clinicopathological factors such as AFP expression, tumor size, differentiation, MVI, and clinical stage. Our ROC analysis showed that MMP-9 CNVs and expression were significant predictors of HCC risk.
The gene copy number can be used as an indicator of major diseases (18). For example, CNV of zinc finger matrine type 4 has the potential as a diagnostic indicator of hematological malignancies (19). A circulating tumor DNA derived CNV detection might be feasible for colorectal cancer (20). Single nucleotide variation of MMP-9 has been studied in breast cancer (21) and gallbladder cancer (22). In this study, we found a higher copy number of MMP-9 in HCC tumor tissues than their adjacent non-tumor tissues.
MMP-9 high expression was found to be correlated to the increase in copy number of MMP-9 in patients with colorectal cancer (18) and brain glioma (23). High expression of MMP-9 is strongly associated with HCC invasion and metastasis (6,24). In this study, the achieved result showed that MMP-9 expression level was higher in tumor tissues than that in adjacent normal tissues, and MMP-9 CNVs appeared only in tumor tissues. The previous study suggested that increased expression levels of the MMP-9 gene were associated with its copy number gains (18). However, our study was unable to find a link between MMP-9 CNV and mRNA expression in tumor tissues, and both MMP-9 copy number gain (16/35) and MMP-9 copy number loss (19/35) were found in our HCC tumor tissues.
MVI is the most relevant risk factor for tumor recurrence in HCC (25). In HCC patients, higher MVI grades and worse differentiation were found to be closely related to the unfavorable prognosis of HCC (3,26). A previous study found that 9p24.2-p21.1 recurrent loss and 8q11.21-q24.3 increase were correlation with the high tumor grade and MVI in HCC patients (27). Here, we found that MMP-9 copy number gains were closely related to tumor size, clinical stage, differential differentiation, and high MVI grading in HCC patients.
Serum AFP is the most widely used serological marker for the diagnosis of HCC (28). AFP was found to induce MMP-9 expression by activating protein kinases and transcription factors (29). Our study found that AFP status in the MMP-9 CNV group was significantly higher than that in non-CNV group. Therefore, we speculate that there is a vital role of AFP in MMP-9 CNV and its expression in HCC patients. Future work is needed to verify our hypothesis.
Strengths and limitations of this study
- MMP-9 CNVs appeared only in tumor tissues.
- MMP-9 CNVs were significantly correlated with AFP, tumor size, differentiation, MVI and clinical stage in the HCC patients.
- ROC curves showed that the CNVs and expression levels of MMP-9 were significant predictors of HCC.
Conclusions
This study showed that copy number gains and higher expression of MMP-9 existed in HCC tumor tissues. MMP-9 CNVs were associated with a series of clinical indicators, including AFP status, tumor size, differentiation, MVI, and clinical stage. Our findings indicate that MMP-9 CNVs have potential diagnostic value for HCC screening and prognosis.
Acknowledgments
The authors are grateful to Chunnian Wang and Xiaolong Lai for their assistance in collecting patients and preparing the manuscript.
Funding: The research was supported by the grants from:
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.11.52). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Ningbo Medical Center Lihuili Hospital (Project Identification Code: DYLL2018028), and informed consent was obtained from all patients before the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet 2012;379:1245-55. [Crossref] [PubMed]
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Imamura H, Matsuyama Y, Tanaka E, et al. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J Hepatol 2003;38:200-7. [Crossref] [PubMed]
- Wang CH, Zhu XD, Ma DN, et al. Flot2 promotes tumor growth and metastasis through modulating cell cycle and inducing epithelial-mesenchymal transition of hepatocellular carcinoma. Am J Cancer Res 2017;7:1068-83. [PubMed]
- Li T, Zhu Y, Han L, et al. VEGFR-1 activation-induced MMP-9-dependent invasion in hepatocellular carcinoma. Future Oncol 2015;11:3143-57. [Crossref] [PubMed]
- Arii S, Mise M, Harada T, et al. Overexpression of matrix metalloproteinase 9 gene in hepatocellular carcinoma with invasive potential. Hepatology 1996;24:316-22. [Crossref] [PubMed]
- Zhang Y, Shen Y, Cao B, et al. Elevated expression levels of androgen receptors and matrix metalloproteinase-2 and -9 in 30 cases of hepatocellular carcinoma compared with adjacent tissues as predictors of cancer invasion and staging. Exp Ther Med 2015;9:905-8. [Crossref] [PubMed]
- Wang L, Ma YT, Xie X, et al. Interaction between MMP-9 gene polymorphisms and smoking in relation to myocardial infarction in a Uighur population. Clin Appl Thromb Hemost 2012;18:72-8. [Crossref] [PubMed]
- Milosevic N, Nikolic N, Djordjevic I, et al. Association of Functional Polymorphisms in Matrix Metalloproteinase-9 and Glutathione S-Transferase T1 Genes with Temporomandibular Disorders. J Oral Facial Pain Headache 2015;29:279-85. [Crossref] [PubMed]
- Sun B, Qiu H, Zhao F, et al. The rs9509 polymorphism of MMP-9 is associated with risk of hemorrhage in brain arteriovenous malformations. J Clin Neurosci 2012;19:1287-90. [Crossref] [PubMed]
- Wang XL, Liu O, Qin YW, et al. Association of the polymorphisms of MMP-9 and TIMP-3 genes with thoracic aortic dissection in Chinese Han population. Acta Pharmacol Sin 2014;35:351-5. [Crossref] [PubMed]
- Nowakowska B. Clinical interpretation of copy number variants in the human genome. J Appl Genet 2017;58:449-57. [Crossref] [PubMed]
- Itsara A, Cooper GM, Baker C, et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet 2009;84:148-61. [Crossref] [PubMed]
- Inaki K, Liu ET. Structural mutations in cancer: mechanistic and functional insights. Trends Genet 2012;28:550-9. [Crossref] [PubMed]
- Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science 2001;291:1304-51. [Crossref] [PubMed]
- Stabley DL, Harris AW, Holbrook J, et al. SMN1 and SMN2 copy numbers in cell lines derived from patients with spinal muscular atrophy as measured by array digital PCR. Mol Genet Genomic Med 2015;3:248-57. [Crossref] [PubMed]
- Baer M, Nilsen TW, Costigan C, et al. Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res 1990;18:97-103. [Crossref] [PubMed]
- Bigagli E, De Filippo C, Castagnini C, et al. DNA copy number alterations, gene expression changes and disease-free survival in patients with colorectal cancer: a 10 year follow-up. Cell Oncol (Dordr) 2016;39:545-58. [Crossref] [PubMed]
- Wan J, Gao Y, Zhao X, et al. The association between the copy-number variations of ZMAT4 and hematological malignancy. Hematology 2011;16:20-3. [Crossref] [PubMed]
- Molparia B, Oliveira G, Wagner JL, et al. A feasibility study of colorectal cancer diagnosis via circulating tumor DNA derived CNV detection. PLoS One 2018;13:e0196826. [Crossref] [PubMed]
- Beeghly-Fadiel A, Lu W, Shu XO, et al. MMP9 polymorphisms and breast cancer risk: a report from the Shanghai Breast Cancer Genetics Study. Breast Cancer Res Treat 2011;126:507-13. [Crossref] [PubMed]
- Sharma KL, Misra S, Kumar A, et al. Higher risk of matrix metalloproteinase (MMP-2, 7, 9) and tissue inhibitor of metalloproteinase (TIMP-2) genetic variants to gallbladder cancer. Liver Int 2012;32:1278-86. [Crossref] [PubMed]
- Xue Q, Cao L, Chen XY, et al. High expression of MMP9 in glioma affects cell proliferation and is associated with patient survival rates. Oncol Lett 2017;13:1325-30. [Crossref] [PubMed]
- Tomizawa M, Shinozaki F, Motoyoshi Y, et al. Niclosamide suppresses migration of hepatocellular carcinoma cells and downregulates matrix metalloproteinase-9 expression. Oncol Lett 2015;10:3515-8. [Crossref] [PubMed]
- Reginelli A, Vanzulli A, Sgrazzutti C, et al. Vascular microinvasion from hepatocellular carcinoma: CT findings and pathologic correlation for the best therapeutic strategies. Med Oncol 2017;34:93. [Crossref] [PubMed]
- Cong WM, Bu H, Chen J, et al. Practice guidelines for the pathological diagnosis of primary liver cancer: 2015 update. World J Gastroenterol 2016;22:9279-87. [Crossref] [PubMed]
- Kim TM, Yim SH, Shin SH, et al. Clinical implication of recurrent copy number alterations in hepatocellular carcinoma and putative oncogenes in recurrent gains on 1q. Int J Cancer 2008;123:2808-15. [Crossref] [PubMed]
- Bai DS, Zhang C, Chen P, et al. The prognostic correlation of AFP level at diagnosis with pathological grade, progression, and survival of patients with hepatocellular carcinoma. Sci Rep 2017;7:12870. [Crossref] [PubMed]
- Konold T, Moore SJ, Bellworthy SJ, et al. Evidence of effective scrapie transmission via colostrum and milk in sheep. BMC Vet Res 2013;9:99. [Crossref] [PubMed]


