
This article has an erratum available at: http://dx.doi.org/10.21037/tcr-2023-09 the article has been update on 2023-12-12 at here.
High-mobility group A1 (HMGA1) gene expressions in various colorectal cancer cell lines and correlation with prognosis
Introduction
The high-mobility group A protein family is characterized by small nuclear proteins with elevated mobility. The HMGA family has four members, including three HMGA1 proteins from alternative splicing, which are HMGA1a, HMGA1b, and HMGA1c, with the fourth protein member being HMGA2. HMGA1 isoforms are located on chromosome 6p21, whereas HMGA2 is transcribed by another gene on chromosome 12q15 (1,2). HMGA1 lacks self-transcriptional activity; thus, it mainly regulates chromatin structures and promotes the interaction between the transcriptional regulatory proteins and downstream DNA, which contains three AT richer domains, known as “AT-hook,” and is also known as an architectural transcription factor.
Previous studies proved that HMGA1 not only promotes tumorigenesis but also enhances the malignant progression of different types of cancer (lung cancer, stomach adenocarcinoma, colorectal cancer, hepatocellular carcinoma, pancreatic adenocarcinoma, kidney carcinoma, bladder urothelial carcinoma, head, and neck squamous cell carcinoma). It was reported that HMGA1 has an important role in promoting thyroid cancer through inhibition of p53 and the induction of TGF-β1 (3), moreover, TGF-β1 induces HMGA1 expression (4). HMGA1 also promotes colorectal cancer development, primarily through transcriptional regulation via the Wnt signaling pathway (5,6), miR-137, and miR-214 (7,8). The rise of metabolomics in recent years has also shown that HMGA1 can increase glucose uptake, promote aerobic glycolysis (9), and promote the development of colorectal cancer (10,11). IL-24 modulates the high mobility group (HMG) A1/miR222/AKT signaling in lung cancer cells (12).
In this study, we aimed to investigate the oncogenic role of HMGA1 in various cancers by analyzing the clinical overall ten-year survival rate and expression level of HMGA1 in the following cancers: head and neck squamous cell carcinoma, lung cancer, stomach and colorectal cancer, liver and pancreatic cancer, cholangiocarcinoma, kidney cancer, and bladder urothelial carcinoma. Our results suggest that HMGA1 has a role in various cancers, and thus targeting this protein family will have beneficial impacts on the survival rate of the cancer patients.
Methods
Cell culture
We obtained DLD1, HCT116, HCT8, LOVO, HT29, SW480, SW620, and RKO cell lines from the American Type Culture Collection, and these were maintained using Dulbecco’s Modified Eagle Medium (DMEM + 10% DMEM medium + 10% fetal bovine serum + 1% penicillin and streptomycin) (GIBCO) at 37 °C in a 50 mL/L CO2 atmosphere.
Public dataset analysis
The cancer expression levels of profiling studies that included matched clinical information were acquired from The Cancer Genome Atlas database. The datasets were classified into two groups: expression datasets in tumor tissues and normal matched adjacent tissues. Using receiver operating characteristic (ROC) curve analysis, combined with sensitivity and specificity chosen as a cutoff point, the expression of HMGA1 in tumor patients was used to create two groups: the lower expression group and the higher expression group. Based on the overall survival time of patients derived from the clinical dataset, we analyzed the difference between the lower and higher HMGA1 expression groups and compared the 10-year overall survival rate by Kaplan-Meier analysis.
mRNA extraction and q-PCR
We used real-time quantitative PCR (QPCR) to analyze the expression of the HMGA1, CBX7, E-cadherin, and β-catenin genes (normalized to the expression of β-actin). Total RNAs were extracted using TRIzol (TIANGEN) according to the manufacturer’s instruction. Total RNA (2 µg) was reverse transcribed using Prime Script RT reagent kit (TaKaRa). The reverse transcription step was 37 °C for 15 min and 95 °C for 5 min. QPCR was performed using SYBR green (Bimake) and the ABI Step One Plus real-time PCR system (Applied Biosystems). We mixed the SYBR Green PCR Master Mix 10 µL with primers 200 nM, cDNA template 1 µL, and deionized water with up to 20 µL of volume. The steps of PCR were 95 °C for 3 min for denaturation, 95 °C for 15 s for annealing, and 60 °C for 30 s for the extension, for 40 cycles. The primer sets used to amplify the gene expression in the study are shown in Table 1.
Table 1
| Gene | Primers |
|---|---|
| β-actin | F: CACAGAGCCTCGCCTTTGCC |
| R: ACCCATGCCCACCATCACG | |
| HMGA1 | F: GCTGGTAGGGAGTCAGAAGGA |
| R: CGAAAGGCCTTCAACTGCAAAT | |
| E-cadherin | F: CGAAAGGCCTTCAACTGCAAAT |
| R: ACTGGTACTTCTTGACATCTG | |
| CBX7 | F: GCGTGCGGAAGGGTAAAGT |
| R: GCTTGGGTTTCGGACCTCTC | |
| β-catenin | F: AAAGCGGCTGTTAGTCACTGG |
| R: CGAGTCATTGCATACTGTCCAT |
Statistical analysis
We used the Statistical Package for the Social Sciences (SPSS version 20.0; IBM New York, NY, USA) for statistical analysis, and P value <0.05 was considered statistically significant.
Results
HMGA1 expression level up-regulated in lung cancer (Figure 1)
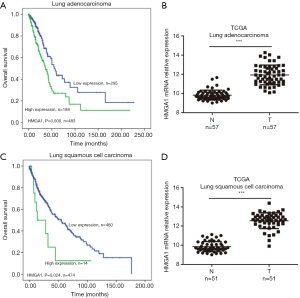
In the TCGA database, we observed two subtypes of lung cancers: lung adenocarcinoma and lung squamous cell carcinoma. HMGA1 was more highly expressed in cancers than in normal adjacent tissues (Figure 1B,D), and its higher expression level predicted worse clinical prognosis (Figure 1A,C), with statistical significance (P<0.05). Surprisingly, we found HMGA1 to be one of the biomarkers in lung cancer, not only because of its elevated expression but also because it was an indicator of the worse overall clinical survival.
HMGA1 expression level up-regulated in the stomach and colorectal cancer (Figure 2)
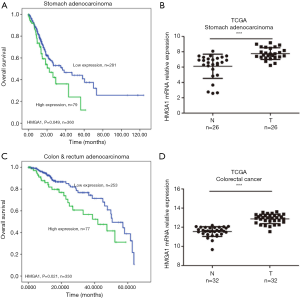
TCGA database showed similar results for the stomach and colorectal cancers. We found higher expression levels of HMGA1 in tumor tissues compared to adjacent normal tissues (Figure 2B,D), and the higher expression group had a significantly worse prognosis for 10-year survival compared to the lower expression group (Figure 2A,C) (P<0.05). A high expression level of HMGA1 was significant and correlated with worse prognosis in the stomach and colorectal cancers.
High expression levels of HMGA1 in the liver and pancreatic cancer (Figure 3)
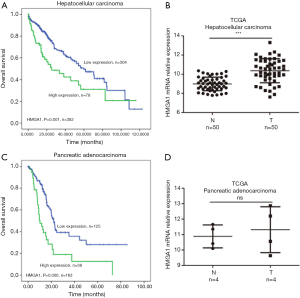
We found the liver and pancreatic cancer patients with elevated levels (prognostic values by Kaplan-Meier analysis) of HMGA1 had a worse prognosis for survival compared to the lower HMGA1 level patients with the same cancers (Figure 3A,C). We also analyzed the HMGA1 expressions in normal control tissues and compared them with liver and pancreatic cancer tissues (Figure 3B,D). The clinical data showed a significant (P<0.05) correlation with HMGA1 expression in cancer (liver and pancreatic) development and prognosis.
High expression levels of HMGA1 in kidney cancer (Figure 4)
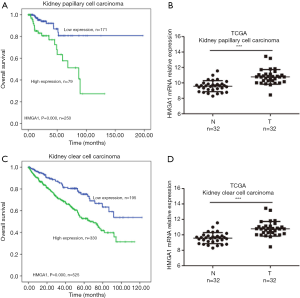
We analyzed the HMGA1 expression level in kidney papillary cell carcinoma and kidney clear cell carcinoma. We found that HMGA1 expression level correlated with the clinical overall survival time of patients. The data showed that the lower expression HMGA1 group had a significantly higher (P<0.05) ten-year survival rate than the high expression group (Figure 4A,C). We also analyzed the expression level of HMGA1 in kidney tumor tissues and matched normal tissues; the results revealed that HMGA1 had a higher expression (significantly, P<0.05) in tumor tissue than in normal tissues (Figure 4B,D). These findings indicate that HMGA1 also serves as a biomarker and can predict clinical overall survival time in kidney cancers.
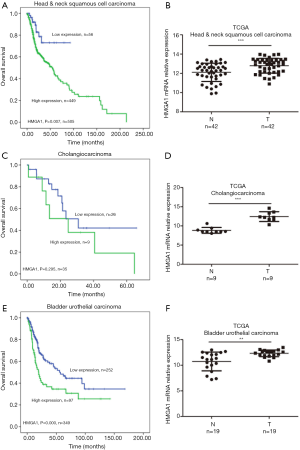
High expression levels of HMGA1 in head and neck squamous cells, cholangio, and bladder urothelial cancers were correlated with worse prognosis
The mRNA values of HMGA1 expression levels of head and neck squamous cell carcinoma, cholangiocarcinoma, and urothelial bladder carcinoma showed similar results as the above. In head and neck squamous cell carcinoma, we found a higher expression level of HMGA1 in tumor tissues compared to normal adjacent tissues. The overall survival time indicated that the higher expression HMGA1 group correlated with shorter survival time (Figure 5A,B). In cholangiocarcinoma, we observed the similar to the above expression level in normal tissues and tumor tissues. The higher expression group of HMGA1 showed worse clinical survival time (Figure 5C,D). In urothelial bladder carcinoma, we found similar results as the above: there was a higher expression level in tumors, which indicated a worse overall clinical survival time (P<0.05) (Figure 5E,F).
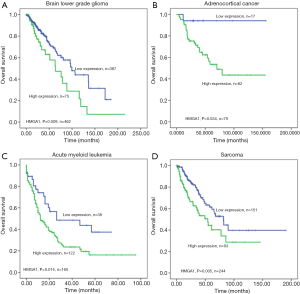
The HMGA1 expression level was correlated with worse overall survival in the following cancers
We found TCGA clinical data related to HMGA1 in different cancers: lower-grade glioma of the brain (Figure 6A), adrenocortical cancer (Figure 6B), acute myeloid leukemia (Figure 6C), and sarcoma (Figure 6D). The analysis of these results differentiated HMGA1 expression level of tumor patients into high and low groups, and the overall survival of the high expression group was significantly lower (P<0.05) compared to the low expression group.
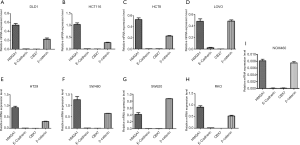
mRNA expression of HMGA1 in different clinical cancer cell lines
The HMGA1 gene is overexpressed in cancer, with higher levels indicating poor prognosis across various tumor types. To further verify that HMGA1 is a tumor marker, we selected eight strains of colorectal cancer cell lines (DLD1, HCT116, HCT8, LOVO, HT29, SW480, SW620, and RKO), a normal human colon mucosal epithelial cell line NCM460 and mRNA expression levels of four genes (HMGA1, E-cadherin, CBX7, and β-catenin) using q-PCR. A large number of studies have reported a low expression of CBX7 and E-cadherin in tumor cells, while in colorectal cancer, β-catenin is highly expressed. We found that in the eight strains of colorectal cancer tumor cell lines, the mRNA level of HMAG1 was significantly higher than that of CBX7 and E-cadherin, and the expression level was similar to that of β-catenin. Although the mRNA level of HMGA1 and β-catenin were higher than that of CBX7 and E-cadherin in NCM460, the relative mRNA level was much lower than that of eight strains of colorectal cancer cell lines. These results show strongly suggest that the level of HMGA1 expression in colorectal cancer is elevated, and HMGA1 is an effective tumor marker (Figure 7).
Discussion
HMGA1 is an oncoprotein that is involved in tumorigenesis and malignant tumor progression. As an oncogene, HMGA1 is upregulated in many different cancers. Huang et al. [2015] revealed that overexpression of HMGA1 correlates with the malignant status and prognosis of breast cancer (13). HMGA1 promotes gastric cancer oncogenic and glycolytic phenotypes by regulating c-myc expression (14). Cheng et al. [2019] demonstrated that HMGA1 exacerbates tumor progression by activating miR-222 through PI3K/Akt/MMP-9 signaling pathway in uveal melanoma (15). In the present study, by analyzing data from the TCGA database, we revealed that HMGA1 is an oncogene in malignant cancers and is highly expressed in malignant cancer patients. The patients with a high expression of HMGA1 showed a shorter overall survival rate compared to lower expression patients in different types of cancers. Recent studies have shown that HMGA1 knockdown or mutation will increase the efficacy of gefitinib in lung cancer cells (16). D’Angelo et al. [2014] reported that HMGA1 proteins lead to chemo-resistance against cetuximab and 5-fluorouracil in colon and thyroid cancer (17). These findings and the present study’s results suggest that the targeting of the HMGA1 protein and a deeper understanding of the underlying mechanisms in tumorigenesis and advanced progression will help in the treatment and management of different types of malignant cancers. Further studies are needed to characterize HMGA1 coordination with downstream functional pathways and correlation with functional proteins.
Conclusions
HMGA1 is an oncofetal gene involved in tumorigenesis and malignant progression. In our study, we revealed that HMGA1 was highly expressed in different cancers, and the high expression level of HMGA1 correlates with worse clinical prognosis. Furthermore, we found a high expression of HMGA1 in lung cancers. With molecular precision treatment becoming the direction of future clinical cancer treatment, our results and the TCGA data suggest that HMGA1-targeted precision therapy can reduce the occurrence of malignant progression of tumors and benefit the survival rate of the patients.
Acknowledgments
Funding: This study was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.10). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sumter TF, Xian L, Huso T, et al. The High Mobility Group A1 (HMGA1) Transcriptome in Cancer and Development. Curr Mol Med 2016;16:353-93. [Crossref] [PubMed]
- Wang Y, Hu L, Zheng Y, et al. HMGA1 in cancer: Cancer classification by location. J Cell Mol Med 2019;23:2293-302. [Crossref] [PubMed]
- Frasca F, Rustighi A, Malaguarnera R, et al. HMGA1 inhibits the function of p53 family members in thyroid cancer cells. Cancer Res 2006;66:2980-9. [Crossref] [PubMed]
- Zhong J, Liu C, Zhang QH, et al. TGF-beta1 induces HMGA1 expression: The role of HMGA1 in thyroid cancer proliferation and invasion. Int J Oncol 2017;50:1567-78. [Crossref] [PubMed]
- Xian L, Georgess D, Huso T, et al. HMGA1 amplifies Wnt signalling and expands the intestinal stem cell compartment and Paneth cell niche. Nat Commun 2017;8:15008. [Crossref] [PubMed]
- Resar L, Chia L, Xian L. Lessons from the Crypt: HMGA1-Amping up Wnt for Stem Cells and Tumor Progression. Cancer Res 2018;78:1890-7. [Crossref] [PubMed]
- Liang L, Li X, Zhang X, et al. MicroRNA-137, an HMGA1 target, suppresses colorectal cancer cell invasion and metastasis in mice by directly targeting FMNL2. Gastroenterology 2013;144:624-35.e4. [Crossref] [PubMed]
- Chandrasekaran KS, Sathyanarayanan A, Karunagaran D. MicroRNA-214 suppresses growth, migration and invasion through a novel target, high mobility group AT-hook 1, in human cervical and colorectal cancer cells. Br J Cancer 2016;115:741-51. [Crossref] [PubMed]
- Chiefari E, Foti DP, Sgarra R, et al. Transcriptional Regulation of Glucose Metabolism: The Emerging Role of the HMGA1 Chromatin Factor. Front Endocrinol (Lausanne) 2018;9:357. [Crossref] [PubMed]
- Williams MD, Xian L, Huso T, et al. Fecal Metabolome in Hmga1 Transgenic Mice with Polyposis: Evidence for Potential Screen for Early Detection of Precursor Lesions in Colorectal Cancer. J Proteome Res 2016;15:4176-87. [Crossref] [PubMed]
- Williams MD, Zhang X, Belton AS, et al. HMGA1 drives metabolic reprogramming of intestinal epithelium during hyperproliferation, polyposis, and colorectal carcinogenesis. J Proteome Res 2015;14:1420-31. [Crossref] [PubMed]
- Panneerselvam J, Srivastava A, Muralidharan R, et al. IL-24 modulates the high mobility group (HMG) A1/miR222 /AKT signaling in lung cancer cells. Oncotarget 2016;7:70247-63. [Crossref] [PubMed]
- Huang R, Huang D, Dai W, et al. Overexpression of HMGA1 correlates with the malignant status and prognosis of breast cancer. Mol Cell Biochem 2015;404:251-7. [Crossref] [PubMed]
- Cao XP, Cao Y, Zhao H, et al. HMGA1 promoting gastric cancer oncogenic and glycolytic phenotypes by regulating c-myc expression. Biochem Biophys Res Commun 2019;516:457-65. [Crossref] [PubMed]
- Cheng Y, Cheng T, Zhao Y, et al. HMGA1 exacerbates tumor progression by activating miR-222 through PI3K/Akt/MMP-9 signaling pathway in uveal melanoma. Cell Signal 2019;63:109386. [Crossref] [PubMed]
- Wang YT, Pan SH, Tsai CF, et al. Phosphoproteomics Reveals HMGA1, a CK2 Substrate, as a Drug-Resistant Target in Non-Small Cell Lung Cancer. Sci Rep 2017;7:44021. [Crossref] [PubMed]
- D'Angelo D, Mussnich P, Rosa R, et al. High mobility group A1 protein expression reduces the sensitivity of colon and thyroid cancer cells to antineoplastic drugs. BMC Cancer 2014;14:851. [Crossref] [PubMed]


