
Integrative analysis of mRNA, miRNA and lncRNA profiles reveals the commonness between bladder cancer and breast cancer
Introduction
Urinary bladder cancer (BLCA) is the ninth most common malignant disease and the thirteenth most common cause of cancer death worldwide (1). Previous studies in BLCA identified signatures associated with stage and outcomes and progression, but the biological and clinical significance of these signatures remain unclear. Therefore, there is an urgent need to identify new biomarkers for a more precise, biology-based approach to the BLCA therapy. Breast cancer (BRCA) is one of the most common cancers with greater than 1,300,000 cases and 450,000 deaths each year worldwide (2). For BRCA, there are some maturely clinical strategies for cure patients owing to widely studies on this cancer. In practice, a more in-depth understanding of the genetic basis of BRCA has led to the development of new treatments and diagnostic tools (2). Recent studies have found that BLCA and BRCA are shared commonness on many areas, such as biological mechanism, molecular subtypes and clinical stage. Three independent research groups identified intrinsic subtypes of muscle invasive BLCA shares molecular signatures with some forms of BRCA (3). Therefore, the analyses of the genetic commonality shared by two cancers are important, and a mature knowledge of BRCA can help highlight the treatment and prognosis of BLCA.
Genetics is already being used to direct clinical decision-making and the contribution of it is likely to increase (4). It is no doubt that combining clinical data and biomarker measurements on a massive scale will do much to improve human health. Expedient biomarkers will be useful in cancer early prediction, diagnostic and decision of treatment. Recently, there are a great many studies on detection and application of tumor biomarker in BLCA and BRCA. Damrauer et al. (5) created a 47-gene predictor (BASE47) that can accurately classify high-grade BLCA into basal-like and luminal type tumors. Cardoso et al. (6) provided prospective evidence of treatment decisions in early-stage BRCA by combining 70-gene signature (MammaPrint) (7) and clinical features, and developed the criteria in selecting patients for adjuvant chemotherapy. These examples clearly demonstrated the power of biomarkers in treatment and prognosis of BRCA and BLCA.
Currently, non-coding RNA (ncRNA) families are being vigorously researched for their physiological and pathological implications (8). MiRNAs are small ncRNAs (~22 nts) and are considered central post-transcriptional gene regulators, which act through transcript degradation and/or translation suppression in the case of mRNAs (9). LncRNA, noncoding transcripts usually longer than 200 nucleotides in length, exhibit numerous functions, many of which are under debate or remain to be uncovered (4). Recent studies suggested that lncRNAs could play a sponge role, competing with other genes for miRNA binding and therefore reduce the regulatory effect of miRNAs on targeted mRNAs (8). This recently proposed post transcriptional mechanism alters the components of endogenous regulatory interaction networks: ncRNAs sharing microRNA response elements (MREs) with mRNAs can act as miRNA sponge and involve in the competing endogenous RNA (ceRNA) activity (8). In the last few years, ceRNA have emerged as an important class of post-transcriptional regulators that alter gene expression through a miRNA-mediated mechanism (9). The majority of biomarker lncRNAs reported by now are derived from studies based on particular type of cancer, for example Second Chromosome Locus Associated With Prostate 1 (SChLAP1) was identified as a prostate cancer—associated lncRNA (10). Some researches indicated that deregulated lncRNAs were implicated in the occurrence and development of BLCA (7). Yang et al. proved that the over expression of miR-346 reduced the expression of SRCIN1 (SRC Kinase Signaling Inhibitor 1) and promoted cell proliferation, colony formation, and sensitivity to Docetaxel (Doc) in BRCA (11). Moreover, Jadaliha et al. used MALAT1 (Metastasis Associated Lung Adenocarcinoma Transcript 1) knockdown/over-expression experiments to confirm the functional significance of MALAT1 as a metastasis driver and a prognostic factor in ER negative, lymph node negative BRCA (12). Furthermore, UCA1 (Urothelial Cancer Associated 1) is a lncRNA reported to promote cell proliferation in both BLCA and BRCA (10,13). Besides, accumulating discoveries of lncRNA functions in various biological processes have revealed the potential of lncRNAs acting as cancer biomarkers. However, to date, there are still little known about a genome-wide expression and function analysis of lncRNAs in BLCA. The molecular mechanisms underlying the ncRNAs role in carcinogenesis and progression of urinary BLCA remain largely unclear (14). Thus, searching lncRNAs signature might be of clinical value for diagnosis and prognosis of BLCA and BRCA. Furthermore, by comparison of similarities between BLCA and BRCA at the molecular level, an abundant BRCA knowledgebase may be utilized to improve the clinical treatment and prognosis of BLCA.
In the present study, we sought to reveal genetic commonness between BLCA and BRCA based on multiple transcriptome profiles including the mRNA, lncRNA and miRNA profiles of BLCA and BRCA patients generated from the Cancer Genomic Atlas (TCGA) project. Functional enrichment analysis based on differential expression genes of BLCA and BRCA was performed to identify cancer-associated annotations and pathways shared by two cancers. By constructing PPI network, we detected the functional organization shared by both cancers, and found some hub mRNA which can play critical role in both cancers’ progress. To further explore the similarity of RNA expression patterns that distinguish tumor from normal patients in BLCA and BRCA, we implemented machine learning analysis to mRNA and lncRNA profiles, and constructed cancer predictor which may help early prediction on both cancers. Moreover, we found some important feature mRNA/lncRNA with high distinguishing ability that may be novel potential biomarkers shared by both cancers. Besides, a comprehensive integrative analysis was applied to construct ceRNA networks of BLCA and BRCA. From these two ceRNA networks, we identified some common cancer-associated ceRNA pairs which may help uncover the relationship between BLCA and BRCA. By these analyses mentioned above, we identified various molecular commonalities of BLCA and BRCA, and these findings indicate that BLCA may be particularly responsive to some treatments for BRCA. The flowchart of our work was shown in Figure 1.
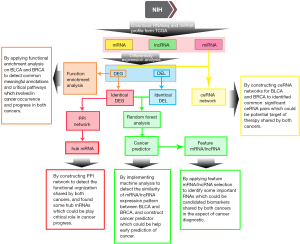
Methods
Genome-wide profiles and data preprocessing
The RNA-seq data and miRNA data of TCGA-BLCA and TCGA-BRCA projects were downloaded from Genomic Data Commons (GDC). The RNA-seq and miRNA datasets of BLCA include 19 primary tumor samples and 19 perineoplastic normal tissue samples from 19 patients. The samples of BRCA in both datasets include 107 tumor samples and 107 perineoplastic normal tissue samples from 107 patients. All of the genetic expression profile were TMM (trimmed mean of M values) (15) normalized and voom (16) transformed by edgeR and limma package of R software (https://www.r-project.org/), respectively.
Differential expression analysis of mRNA and lncRNA
After performed the data preprocessing, differential expression analysis was implemented on RNA-seq data to screen out significant RNAs by limma package of R software. We screen out the differentially expressed RNAs using the following criteria: (I) significance level: p<0.05; (II) |fold change|>1.5; (III) false discovery rate (FDR) <0.05. Then, we extracted differential expressed mRNAs (DEGs) and differential expressed lncRNAs (DELs) from these significant RNAs.
Functional enrichment analysis based on DEGs
In order to explore meaningful annotations of genes and gene products in BLCA and BRCA, gene ontology (GO) enrichment analysis was implemented on RNA expression of both cancers. Because the functions of most lncRNAs were poorly defined, we only used DEGs of two cancers to implement this analysis. As we know, lncRNAs could regulate mRNAs by some biology mechanisms, such as cis-regulation and trans-regulation (9). Therefore, lncRNAs execute functions that might be reflected in related mRNAs. GO enrichment analysis of DEGs can reveal the role of obviously differentially regulated lncRNAs. The ontology has covered domains of biological processes, cellular components and molecular functions. We also performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis on DEGs of two cancers to harvest pathway clusters covering our knowledge on the molecular interaction and reaction networks in differentially regulated gene profiling. DAVID database (http://david.abcc.ncifcrf.gov/) was used for implementing the functional enrichment analysis.
Construction of protein-protein interaction (PPI) network and screening of hub genes
PPI network is the basic method for systematically displaying PPI information. It provides a valuable framework for a better understanding of functional organization. We used identical DEGs shared by both cancers to construct PPI network. The STRING database (http://string.org) was used to get the protein-protein pairs. Then, the Cytoscape software (version 3.4.0; http://cytoscape.org/) was used to visualize the PPI networks and to further explore the association between genes. Subsequently, the k-core scoring was used to determine core mRNA of PPI networks. A higher k-core score means a more central location of an node within a network (17). The MCODE plug-in of Cytoscape was used to perform this analysis.
Construction of cancer predictor based on machine learning method
We next investigate whether the expression of RNA (mRNA and lncRNA) patterns that distinguish normal from tumor samples in BLCA is similar to that of in BRCA by applying machine learning approaches which have been applied widely to cancer prognosis and prediction (18). Specially, we sought to generate gene set predictors that can accurately classify tumor and normal samples. We generated BLCA-type and BRCA-type classifiers with machine learning method based on mRNA and lncRNA profilers of both cancers. We selected DEGs and DELs overlapped in BLCA and BRCA, and then applied random forests (RF) method to construct tumor predictable classifiers: mRNA-type classifier and lncRNA-type classifier. RF is an ensemble classifier that consists of many decision trees and each tree depends on the values of a random vector sampled independently (19). The out-of-bag (OOB) error was used as a value of self-validation and assessed the performance of classifiers. We then used the classifier trained by one cancer to predict the other cancer’s profile. We supposed that the prediction accuracy of classifiers will be nice if the expression patterns of both cancers are similar, and it demonstrated that BLCA and BRCA are similar in terms of oncogenic pathway. The program was implemented with the RF package of R software.
Identification of feature mRNA/lncRNA shared by both cancers
When we got a good cancer predictor, we want to know which feature (mRNA and lncRNA) can make great contributions on identifying tumor samples. In RF method, suppose a tumor-related feature gene G is a good predictor, then it will appear in a large number of split trees. We used Mean Decrease Gini (MDG) to evaluate whether gene G is a feature gene or not. MDG provided possible ways to quantify which genes contribute most to classification accuracy. Greater MDG will indicate that the degree of impurity arising from category could be reduced farthest by gene G, and thus suggests an important feature gene. We sought to identify some feature mRNA and lncRNA which may play an important role in BLCA and BRCA diagnostic. We implemented this analysis with RF package of R software.
Constructing ceRNAs network
The ceRNA hypothesis and many studies demonstrated that all types of RNA transcripts can bind with miRNA through microRNA response elements MREs, and acting as ceRNAs (8,9). In this part, we constructed ceRNAs networks using DEGs, DELs and miRNA profiles of BLCA and BRCA. There were three criteria (20) were used to determine the competing endogenous interactions between lncRNA-mRNA pairs: (I) the lncRNA and mRNA must share significant number of miRNAs. (II) Expression of lncRNA and mRNA must be positively correlated. (III) Those common miRNAs should play similar roles in regulating the expression of lncRNA and mRNA. Hypergenometric test was performed to test whether a lncRNA and mRNA share many miRNAs significantly (significance level: P<0.01) based on starBase v2.0. Pearson correlation coefficient was used to measure the strength of a linear association between two RNAs. As we know, miRNAs are negative regulators of gene expression. If more common miRNAs are occupied by a lncRNA, less of them will bind to the target mRNA, thus increasing the expression level of mRNA. So, the expression of the lncRNA and mRNA in a ceRNA pair should be positively correlated (significance level: P<0.05). The regulation similarity score was defined to check the similarity between miRNAs-lncRNA expression correlation and miRNAs-mRNA expression correlation. We further selected lncRNA-mRNA pairs that regulation similarity score is not equal to zero. The regulation similarity score (20) was defined as below:
regulation similarity score=
where M is the total number of shared miRNAs, k is the k-th shared miRNAs, corr(mk, l) and corr(mk,g) represents the Pearson correlation between the k-th miRNA and lncRNA, the k-th miRNA and mRNA, respectively. The ceRNA networks were constructed by GDCRNATools package of R, and visualized by Cytoscape.
Results
Overview of DEGs and DELs
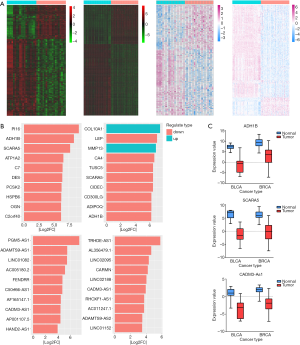
By applying differential expression analysis, a total of 2,149 DEGs (659 mRNAs were up-regulated and 1,490 mRNAs were down-regulated) and 119 DELs (48 lncRNAs were up-regulated and 71 lncRNAs were down-regulated) were identified from RNA-seq data of BLCA, while 2,693 DEGs (953 mRNAs were up-regulated and 1,740 mRNA were down-regulated) and 211 DELs (35 lncRNAs were up-regulated and 176 lncRNA were down-regulated) from RNA-seq data of BRCA (Figure 2A). We displayed the top 10-fold change DEGs and DELs for both cancers in the Figure 2B. Among these biomarkers, Peptidase Inhibitor 16 (PI16) and PGM5 Antisense RNA 1 (PGM5-AS1) were the most down-regulated mRNA and lncRNA in BLCA compared with the adjacent noncancerous tissue, respectively. On the contrary, Collagen Type X Alpha 1 Chain (COL10A1) and TRHDE Antisense RNA 1 (TRHDE-AS1) were the most up-regulated mRNA and the most down-regulated lncRNA in BRCA (Figure 2B). Furthermore, we found that mRNAs Alcohol Dehydrogenase 1B (ADH1B) and Scavenger Receptor Class A Member 5 (SCARA5) as well as lncRNA CADM3 Antisense RNA 1 (CADM3-AS1) were significantly down-regulated in both cancers (Figure 2C).
GO annotations and KEGG pathways of both cancers
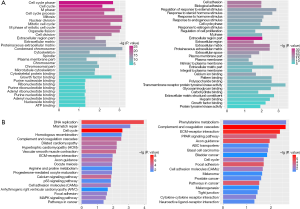
GO enrichment analysis revealed that DEGs in BLCA mostly consisted of cell cycle related terms as these processes were necessary to BLCA cell proliferation (Figure 3A left). These DEGs of BLCA were enriched on extracellular matrix (ECM) related terms (Figure 3A left). ECM is defined as a complex mixture of various proteins that provides structural and mechanical support for cells and tissues, and has an important role in the regulation of gene expression, cell division, survival, shape, and movement (11). Previously study reported that ECM molecules play an important role in the development of invasion, progression and metastasis in BLCA (11). From the GO enrichment analysis of BRCA (Figure 3A right), we found that many DEGs were significantly involved in cell adhesion and biological adhesion. It is now clear that adhesive interactions play a critical role in the process of metastatic tumor dissemination (21,22). Furthermore, these DEGs of BRCA are associated to response to a series of biomacromolecule stimulus, such as steroid hormone and estrogen stimulus. An interesting observation was that these DEGs of BRCA were enriched on ECM related terms, similar to the function enrichment result of BLCA. Some researches indicated that ECM may be involved in various processes of breast tumors growth (23). The DEGs of both cancers were also enriched on many cancer-related KEGG pathways, and we listed significant KEGG pathways in Figure 3B. From the results we found some important pathways shared by both cancers including Cell Cycle, Complement and coagulation cascades, ECM-receptor interaction, Axon guidance, Cell adhesion molecules, Focal adhesion and Pathway in cancer. These KEGG pathways may play an important role in cancer occurrence or progress. For example, Gordon et al. found that altered ECM and specific components of ECM can alter gene expression, cellular proliferation, and invasiveness of the overlying urothelial cells (24). For BRCA, ECM in necessary for normal functional differentiation of mammary epithelia (25). Focal adhesion kinase (FAK) is an important symbolic component that is activated by many stimuli, acting as a biosensor or an integrator to control cell motility (26). FAK is an also important regulator of BLCA cell invasion and migration, and it may become a potential therapeutic target (27). For BRCA, FAK may play a critical role in the cancer by regulating breast stem cells and BRCA stem cells (26,28). Emerging data suggest that FAK can be an effective therapeutic target in breast tumors, particularly in highly invasive triple-negative BRCA (26). Interestingly, we found that some DEGs of BRCA were enriched on pathway of bladder cancer.
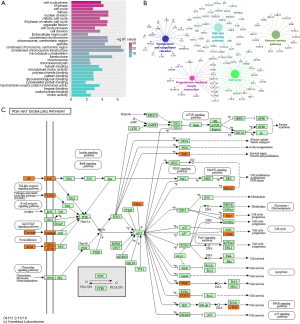
To further explore the commonness between BLCA and BRCA, we selected 1,176 overlapping DEGs shared by two cancers to implement GO and KEGG enrichment analysis. The result showed that these DEGs were significantly enriched on cell cycle related terms, such as cell cycle phase, mitotic cell cycle, cell cycle process and so on, which are necessary to tumor cell proliferation (Figure 4A). The KEGG enrichment analysis revealed that overlapping DEGs of both cancers were enriched on the pathways of cancers significantly. These DEGs were mainly enriched on five kinds of pathways: complement and coagulation cascade, PI3K-Akt signaling pathway, Fanconi anemia pathway, progesterone-mediated oocyte maturation and cell cycle (Figure 4B). In these pathways, PI3K-Akt signaling pathway covered the most genes and was the most affected pathway in BLCA and BRCA (Figure 4C). In recent years, it has been shown that PI3K-Akt signaling pathway, involved in the above and other processes, are frequently disturbed in many human cancers (20). The major functions of the PI3K-Akt signal pathway are to promote growth-factor-mediated cell growth, proliferation, migration and survival (12). Moreover, PI3K-Akt signal pathway plays role not only in tumor development but also in the tumor’s potential response to cancer treatment (20). Thus, many of the new “targeted agents” have been specifically designed to act on PI3K-Akt related targets. Jadaliha et al. found that inhibition of the Akt signaling pathway may improve the efficacy of endocrine therapy for BRCA (12). We thus believe that such endocrine therapy based on PI3K-Akt signaling pathway may help to treat BLCA. Recently, many of the effects of the new targeted anticancer drugs have mechanistic connections with the PI3K-Akt pathway, and therefore a better understanding of this essential crossroad can help to fully exploit the potential benefits of these new target therapies in both cancers.
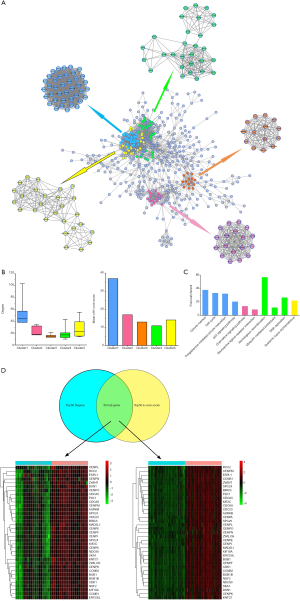
PPI network and key hub gene
To further investigate the similarity function of genes at the protein level in both cancers, we used identical DEGs of both cancers to construct PPI network. The PPI network contained 480 nodes and 2,968 edges (Figure 5A). According to the k-core score and the connection between nodes we got five main sub-networks (five clusters: blue, green, orange, yellow, pink) from the PPI network, and the k-core score of mRNA containing in these sub-networks were shown in the Table S1. We can see that the mRNAs which constitute blue sub-network have the highest degree and k-core scores, and this indicated that the blue sub-network could play critical roles in the carcinogenic process in both cancers (Figure 5B). By applying KEGG enrichment analysis on these sub-networks we found that these sub-networks are enriched on severe signaling pathways, including p53 signaling pathway, chemokine signaling pathway, homologous mediated proteolysis and systemic lupus erythematosus (Figure 5C). Moreover, by taking intersection from genes with top 50 highest degree and top 50 highest k-core score we found 35 hub genes which could be potential biomarkers (Figure 5D). Among these genes, Cyclin Dependent Kinase 1 (CDK1) has highest k-core score (k-core score =37) and highest degree (degree =103). CDK1 is a universal master kinase and is required for the regulation of mitosis (29). Nakayama et al. (10) found that CDK1 plays a critical role in pacilitaxel-induced cell death, and its activity can be a predictor of paclitaxel sensitivity in BRCA treatment. Furthermore, there are other important genes were found, such as Cyclin B1 (CCNB1), Cell Division Cycle 20 (CDC20), BUB1 Mitotic Checkpoint Serine/Threonine Kinase (BUB1) and so on (Figure 5D). The high-expression of CCNB1 can cause uncontrolled growth of the tumor. A study has shown that CCNB1 promotes cancer invasion and metastasis by enhancing epithelial to mesenchymal transition process (30). Moreover, the over-expression of CDC20 is associated with poor prognosis of BLCA (31). In the case of BRCA, CDC20 is a biomarker of triple-negative BRCA and its high expression is associated with high risk of death (32). Besides, over-expression of BUB1 may be a new hallmark for estimating the biological characteristics of BLCA (33). Interestingly, the expression level of BUB1 is associated with different clinical outcomes in BRCA patients and could be a potential therapeutic target (34). In addition, all of these hub genes were contained in blue sub-network. The study of these genes may help develop new means to treat BLCA and BRCA.
Cancer predictors based on RF methods
To further identify the potential relationships between BLCA and BRCA, we analyzed DEGs and DELs shared by two cancers using RF method. We used RNA expression profile of one cancer to train RF model, and applied this model to predict the other cancer samples based on its RNA expression profile. Firstly, we selected 1176 overlapping DEGs and 41 overlapping DELs of both cancers, and we got four datasets: overlapping mRNA expression profile of BLCA (1,176 mRNAs), overlapping mRNA expression profile of BRCA (1,176 mRNAs), overlapping lncRNA expression profile of BLCA (41 lncRNAs) and overlapping lncRNA expression profile of BRCA (41 lncRNAs) respectively. We named these four datasets as mBLCA, mBRCA, lncBLCA and lncBRCA datasets, respectively. Secondly, we used these datasets to construct four types RF classifiers (named as mBLCA classifier, mBRCA classifier, lncBLCA classifier and lncBRCA classifier, respectively). Finally, we applied these classifiers to perform the prediction. The pipeline of analysis and the result were shown in the Figure 6A.
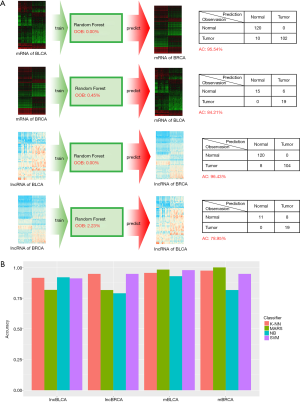
As shown in Figure 6A, all of four type classifiers have low OOB errors. The OOB of mBLCA classifier, mBRCA classifier, lncBLCA classifier and lncBRCA classifier was 0%, 0.45%, 0%, 2.23%, respectively. Interestingly, when we used the classifier constructed by one cancer expression profile to predict the other cancer, the predictor displayed the good predict performance. The classification accuracy for mBLCA, mBRCA, lncBLCA and lncBRCA classifiers were up to 95.54%, 84.21%, 96.43% and 78.95%, respectively. The good predict performance indicating that the mRNA and lncRNA expression patterns that distinguish normal and tumor tissue of BLCA reflect the similarly expression patterns that defined the BRCA. To further validate this result, we additionally used another four classifier algorithms including K-Nearest Neighbor (KNN), Multivariate adaptive regression splines (MARS), Naïve Bayes (NB) and Support Vector Machine (SVM) to observe their predict accuracies. The result showed that all of four classifier algorithms have good performances (Figure 6B). Taken together, our study demonstrated that the RNA expression patterns that distinguish normal from tumor bladder (breast) samples reflect the RNA expression patterns that define the breast (bladder) tumor, which indicated the commonness of both cancers in the aspect of oncogenes’ expression.
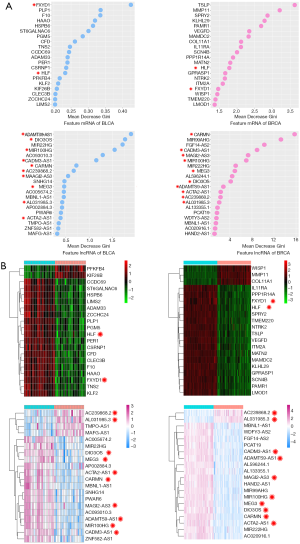
Feature mRNA and lncRNA shared by both cancers
In above section, we constructed four types of cancer predictor with good performance by RF method. We used MDG to measure the distinguishing ability of features (mRNA/lncRNA). We listed the top 20 mRNA/lncRNA according to their MDG on Figure 7A. Their expression heatmaps were shown in Figure 7B. In this analysis, we found 2 mRNA and 10 lncRNA were overlapped in both cancers (RNA with red star in Figure 7). These RNAs play critical roles in tumorigenesis and make contribution to predict BLCA and BRCA tumors. For instance, the FXYD proteins have been proposed to function as regulators of Na, K-ATPase function by lowering affinities of the system for potassium and sodium. Floyd et al. (15) found that FXYD Domain Containing Ion Transport Regulator 1 (FXYD1) low expressed in bladder and breast tumors. Maternally Expressed Gene 3 (MEG3) is an imprinted gene that encodes lncRNA associated with carcinogenicity. Ying et al. (18) suggested that down-regulated MEG3 activates autophagy and induce cell proliferation in BLCA. Sun et al. (35) demonstrated that down-regulation of MEG3 in BRCA tissues affect BRCA cells’ malignant behaviors. These evidences indicate MEG3 is a potential therapeutic target for BLCA and BRCA. The feature RNAs we found in this analysis might shed light on the development of new RNA-based therapeutic target for treating BLCA and BRCA.
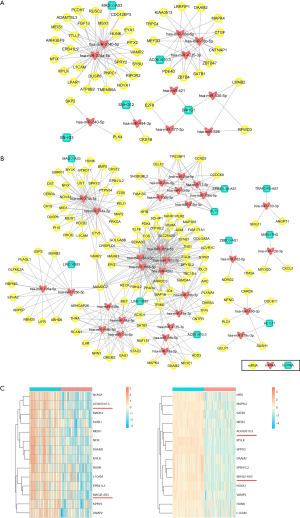
ceRNAs network
According to ceRNA hypothesis, ceRNAs members can compete for the same MREs to regulate each other (8). Under the three criteria mentioned above, we constructed ceRNA networks of BLCA and BRCA base on our microarray data. In this analysis, we found 49 lncRNA-miRNA-mRNA triples combined by 44 DEGs, 5 DELs and 11 miRNAs in ceRNA of BLCA (Figure 8A). For BRCA, we identified 298 lncRNA-miRNA-mRNA triples combined by 113 DEGs, 10 DELs and 30 miRNAs (Figure 8B). These ceRNA pairs may provide a new potential therapeutic possibility for BLCA and BRCA. Interestingly, we found MAGI2 Antisense RNA 3 (MAGI2-AS3) and AC093010.3 were overlapped lncRNAs involved in two cancers-related ceRNA networks, and some common mRNAs were regulated by these two lncRNAs. The heatmaps of these overlapping mRNA/lncRNA expression profiles in both cancers were shown in Figure 8C.
In two cancers-related ceRNA networks, we found that the Membrane- associated Guanylate Kinase, WW and PDZ Domain-containing 2 (MAGI2) acted as hsa-miR-374a-5p and hsa-miR-374b-5p sponge to weaken the inhibition of Myosin Light Chain Kinase (MYLK) expression in both cancers. MYLK is a member of the immunoglobulin gene super family, and has been linked to the proliferative ability of BRCA via extracellular signal-regulated kinase and the P38 pathway; moreover, it is associated with modulation of tumor invasiveness and metastasis in BRCA (36). Robert T. Lawrence et al. (37) indicated that MYLK was involved in immunity and metastasis, and was expressed at higher levels in TNBC. van't Veer et al. (7) identified a circular RNA derived from MYLK gene, termed circRNA-MYLK, as a ceRNAs promotes BLCA progression through modulating VEGFA/VEGFR2 signaling pathway. Many studies found that circRNA-MYLK is a candidate oncogene in BLCA and correlate with BLCA progression. However, little was known of the lncRNA-MYLK in BLCA. In our study, MAGI2-AS3 was constructed ceRNA pairs with MYLK, and it is an interesting candidate in BRCA and BLCA prognostic, which should be validated by relative clinical trials in the future.
The Myeloid Ecotropic Viral Integration Site 1 (MEIS1) transcription factor gene is known to play a crucial role in normal and tumor development (38). We identified that MAGI2-AS3 acted as miRNA sponge to absorbed miRNAs and weakened the inhibition of MEIS1 expression. Beukers et al. (39) indicated that MEIS1 was a target of Polycomb genes in BLCA. In BRCA, MEIS1 may be useful member of a panel of favorable prognostic and predictive markers for BRCA and an understanding of their function may provide useful information about this disease (40). Therefore, we suggested that the ceRNA pair of MAGI2-AS3 and MEIS1 could be a candidate therapeutic target of both cancers. L1 Cell Adhesion Molecule (L1CAM), a transmembrane cell adhesion molecule of the Ig super-family, plays an important role in the development of the nervous system and in the malignancy of human tumors (41). L1CAM is over expressed in many human carcinomas and augments cell motility, invasion and metastasis formation (41,42). We found that lncRNA MAGI2-AS3 also acted as miRNA sponge to weaken the inhibition of L1CAM expression in BLCA and BRCA, they were bound by hsa-miR-374a-5p and hsa-miR-374b-5p. Doberstein et al. (43) found that high expression of L1CAM in primary breast tumors have the worst clinical outcome, which indicated that L1CAM expression could be causally related to the bad prognosis of TNBCs. Faltas et al. (42) suggested that alterations in L1CAM and integrin signaling pathways potentially play a key role in chemotherapy resistance in urothelial carcinoma. It is thus suggested that further study of the functional role of MAGI2-AS3-L1CAM ceRNA pair in BLCA and BRCA could lead to a potential strategy for targeting therapy. Furthermore, the lncRNA AC093010.3 could serve as a sponge for hsa-miR-4735-3p, hsa-miR-18a-5p and hsa-miR-18b-5p to abolish the endogenous suppressive effect on Special AT-rich Binding Protein 1 (SATB1) expression in both cancers. The T cell-enriched transcription factor SATB1 is a nuclear protein which has a high level of expression in thymocytes, and it is an independent prognostic factor and a potential therapeutic target in human cancers (44). Moreover, altering SATB1 regulated the cell migration, invasion and proliferation, along with the cell cycle (45). In BLCA, SATB1 plays a critical role in the tumor progression by regulating genes controlling EMT processes, which makes SATB1 therapeutic target candidate for BLCA (45). For BRCA, some studies observed that the expression level of SATB1 was higher in malignant compared with normal breast tissue, and SATB1 over expression associated with TNM stage, tumor grade, and shorter overall survival (46). In fact, the SATB1 protein has been proposed as a target for BRCA progression and metastasis (44,46). Han et al. (44) suggested that SATB1 protein is expressed during carcinogenesis to alter the gene expression profile of BRCA cells to support an aggressive cell phenotype that promotes tumor growth and metastasis. Thus, AC093010.3-SATB1 pair is a therapeutic target candidate for BLCA and BRCA. These findings suggested that the constructed ceRNA networks of BLCA and BRCA can provide different angle of view for detecting commonness of different types of cancers.
Discussion
The conventional view of gene regulation focused on protein-coding genes until the discovery of numerous miRNAs and lncRNAs. Especially, some researches of deregulated lncRNA expression covering various kinds of cancer indicated that abnormal lncRNA expression might contribute to carcinogenic and progression (47). BLCA and BRCA are highly heterogeneous tumors with variable clinical courses. For BLCA, there are some conventional clinical therapeutic strategies but patients got limited benefit from their (48). Therefore, there is an urgent need to develop biology-based approach to the management of BLCA. On the contrary, for BRCA, there are quit maturely clinical strategies for cure patients owing to deep studies on BRCA (2). Recent studies found BLCA are remarkably similar to BRCA in molecular level, which means that uncover their similarities at the molecular level may be possible to leverage the larger BRCA knowledge base to enable more rapid progress in the clinical management of BLCA (3,5).
In the present study, we used mRNA, miRNA and lncRNA express profiles to explore the commonness between BLCA and BRCA from four perspectives: functional enrichment, PPI network, machine learning and ceRNA network. Our research identified some significantly function annotations shared by both cancers, such as cell proliferation related terms and ECM related terms. Besides these terms mentioned above, we found that the DEGs of BLCA were significantly involved in calcium signaling pathway whereas DEGs of BRCA take part in calcium ion binding. Ionized calcium (Ca2+) as an intracellular second messenger is ubiquitous and controls various critical biological processes (49). The S100 family of proteins, belonging to Ca2+-modulated proteins, is a large group of low molecular weight Ca2+-binding protein (50). Some studies confirmed that S100 Calcium Binding Protein P (S100P) can mediate BLCA tumor growth, metastasis and invasion through the binding of Ca2+ ions, which indicated that S100P can serve as the potentially diagnostic marker and therapy target for BLCA (51). For BRCA, some studies reported that the S100P Ca2+-binding protein could be used as a hallmark to differentiate lesions at high risk of malignant evolution (50). Further investigation into the physiological role of S100 proteins in the mammary gland may lead to the characterization of potentially new therapeutic targets for BRCA. Therefore, an improved knowledge of the functioning of Ca2+ signaling will help in diagnosing and treating cancers. By applying KEGG enrichment analysis we observed that the most overlapping DEGs of both cancers were significantly involved in PI3K-Akt signaling pathway. Meanwhile, some critical oncogenes of BLCA and BRCA take part in the pathway, such as BRCA1 (BRCA1, DNA Repair Associated), BRCA2 (BRCA2, DNA Repair Associated), BUB1B (BUB1 Mitotic Checkpoint Serine/Threonine Kinase B), MYC (MYC Proto-Oncogene, BHLH Transcription Factor) and so on. It is suggested that explore the mechanisms of these alteration carcinogenic pathway is extraordinary helpful in development clinical strategies of early diagnosis and target therapy.
Different types of cancers can be shared similar basic regulatory system, even if the phenotype and tissue origin may differ. To detect the similarity of mRNA interaction relationships, we implemented the PPI network analysis using identical DEGs. We identified hub genes which could play important role in cancer progress. Besides, we also found some other important genes in sub-network. For instance, we found that BRCA1 and BRCA2 with high degree in blue sub-networks. BRCA1 and BRCA2 were widely reported to be BRCA -related genes, and were high risk family of oncogenes (13). In the practice, it is suggested that BRCA1 expression could be used for predict the efficacy of cisplatin-based neoadjuvant chemotherapy and may help to customize therapy in BLCA patients (16).
To further detect the relationships between BLCA and BRCA in the view of expression patterns, we used machine learning methods to analyze identical DEGs and DELs of both cancers. For both cancers, we applied RF algorithms to construct mRNA type classifiers and lncRNA type classifiers, and we found all of four type classifiers have good performances. When applied four additionally classification algorithms including K-NN, MARS, NB and SVM, we got the similar results. These results supported that BLCA and BRCA have similar RNA expression patterns, which indicated that these two cancers may have potential relationships in terms of RNA expression phenotype. Moreover, when constructed RF classifiers we obtained some critical feature mRNAs and lncRNAs with high MDG including FXYD1, MEG3 and so on, which might be candidate biomarkers for diagnostic and treatment.
Importantly, it is known that identifying a smallest possible but most informative subset of genes is the goal of gene selection. A small subset of genes is also desirable in developing gene expression-based diagnostic tools. Although there are some predictor which can accuracy classify cancer subtypes by small number of gene, such as BASE47 (5) for high-grade BLCA and PAM50 (52) for BRCA, there is little research on cancer predictor with small number of lncRNA. For this, we add another analysis to construct mRNA and lncRNA type predictor with smallest number but highest prediction accuracy of RNA subset. We applied multiple support vector machine recursive feature elimination (MSVM-RFE) (53) method to select smallest but most informative subset of RNAs, and constructed cancer classifiers for BLCA and BRCA. Specially, we performed MSVM-RFE on the whole differential expression RNAs including 2149 DEGs of BLCA, 119 DELs of BLCA, 2693 DEGs of BRCA and 211 DELs of BRCA on both cancers. We computed feature ranking score by MSVM-RFE method on BLCA and BRCA. The top 20 ranked RNAs were shown in Figure S1A. The feature subset was examined by 10-fold cross validation of linear SVM classifier trained with features in the subset as input variables. We test feature subsets with size ranging from 1 to 200, and plot the average test errors (generate from 10-fold cross validation) of linear SVM classifiers on gene subsets selected by MSVM-RFE on two cancers (Figure S1B). We take the feature subset with the least average test error as the best feature subset. The best features number of mRNA of BLCA, mRNA of BRCA, lncRNA of BLCA and lncRNA of BRCA were 12, 17, 91 and 5 respectively. Additionally, we constructed four type of predictor, including NB, SVM, K-NN and Artificial Neural Network (ANN). We used ten times leave-one-out method to assess the classification accuracy rate of these different machine learning methods, and the results were shown in Table S2. Interestingly, the predictors constructed by best RNA subset showed good classification accuracy.
Furthermore, the BLCA and BRCA type of ceRNA networks were constructed by applying integration analysis based on mRNA, miRNA and lncRNA profiles. In two cancers-related ceRNA networks, we identified some meaningful ceRNA pairs. For instance, the lncRNA MAGI2-AS3 acted as miRNAs sponge to weaken the inhibition of some critical oncogenes, such as MYLK, MEIS1 and L1CAM in both cancers. The lncRNA AC093010.3 acting as competitor in ceRNA networks of both cancers can absorb miRNAs and regulate SATB1 expression. Besides overlapping ceRNA pairs mentioned above, we found some other critical competitor relationships in each cancer. For instance, the small nucleolar RNA host gene, including Small Nucleolar RNA Host Gene 1 (SNHG1), SNHG3 and SNHG12, could serve as a sponge for miRNA to abolish the endogenous suppressive effect on target mRNAs expression in BLCA ceRNA network. Among these lncRNAs, SNHG1 has been considered as indicator of several human cancers (54), and in this study we found that SNHG1 is ceRNA of hsa-miR-421 and hsa-miR-377-3p target E2F Transcription Factor 8 (E2F8). Some studies demonstrated that deregulation of E2F family activity was correlated with aberrant cell proliferation and in some instances cell death (55). The lncRNA SNHG3 acted as miRNA sponge to absorb miRNAs and weakened the inhibition of S-Phase Kinase Associated Protein 2 (SKP2) expression. Accumulating evidence suggested that SKP2 is an important oncoprotein and a potential target of Wnt regulation in several human cancer cells (56). It is noted that some studies indicated that over expression of SKP2 has been implicated in cell transformation and carcinogenic in BRCA (57). Moreover, in the case of BRCA, the lncRNA Long Intergenic Non-Protein Coding RNA 667 (LINC00667) was a hub node with the highest degree in the BRCA ceRNA network, indicating that it can regulate a great number of mRNAs by competitively bound with miRNAs. For instance, LINC00667 acted as hsa-miR-19a-3p and hsa-miR-19b-3p sponge to weaken the inhibition of Transferrin Pseudogene 1 (TFP1) expression, and acted as hsa-miR-081a-5p, hsa-miR-081b-5p, hsa-miR-081c-5p and hsa-miR-081as-5p sponge to regulate Reversion Inducing Cysteine Rich Protein with Kazal Motifs (RECK) and Inhibitor of DNA Binding 4 (ID4) expressions. Among these mRNAs, Zhang et al. (58) indicated that RECK is an important molecule in preventing tumor cell metastasis in BRCA. RECK is a novel predictor of breast carcinoma for that LINC00667-RECK could emerge as an attractive new drug target in the treatment of BRCA. Some researches indicated that down regulation of Tissue Factor Pathway Inhibitor (TFPI) induced intracellular tyrosine signaling and increased the self-sustained growth and metastatic abilities of BRCA cell lines (59). In addition, ID4 is a potential tumor suppressive gene that involve in aggressiveness of tumor behavior (60). Thus, LINC00667-TFPI and LINC00667-ID4 pair might be proven useful as a genetic marker to predict early metastasis and progress. Furthermore, we found that Long Intergenic Non-Protein Coding RNA 893 (LINC00893) acted as ceRNA in the BRCA ceRNA network, and it regulated the expression of 11 mRNAs, such as Ubiquitin Specific Peptidase 2 (USP2) and EPH Receptor A2 (EPHA2). Moreover, the lncRNA Pvt1 Oncogene (PVT1), lncRNA EPB41L4A Antisense RNA 1 (EPB41L4A-AS1) were ceRNA of seven miRNAs targeting Nuclear Factor I B (NFIB), Family With Sequence Similarity 13 Member C (FAM13C), SH3 Domain Binding Glutamate Rich Protein Like 2 (SH3BGRL2), CUGBP Elav-Like Family Member 2 (CELF2), Tumor Protein P53 Inducible Nuclear Protein 1 (TP53INP1), Cyclin D2 (CCND2) and Coiled-Coil Domain Containing 68 (CCDC68). LncRNA Zinc Finger BED-Type Containing 3 (ZBED3-AS1) was ceRNA of hsa-miR-382-5p targeting NDRG Family Member 2 (NDRG2) and Integral Membrane Protein 2A (ITM2A). These RNA interactions can supply novel perspective for the tumorigenes of BLCA and BRCA.
In summary, we uncovered molecular commonness between BLCA and BRCA by applying a comprehensive integrative analysis. Our results suggested that specific genes, lncRNAs and ceRNA pairs could be valuable for diagnosis and therapy of cancers and be of biological importance. Our data might lay a foundation for further functional research of commonness between BLCA and BRCA. Furthermore, this computational framework can be easily extended to other cancer types or diseases, if samples are simultaneously measured from mRNA, lncRNA and miRNA expression levels.
Some limitations of this study should be pointed out. On one hand, the increasing volume of multi-omics data can help improve the accuracy and stability of machine learning analysis, and integrate more molecular biomarkers to explore the relationship between BRCA and BLCA are our future work. On the other hand, although our study provided biological insights into the molecular similarity of BLCA and BRCA, additional experiments will be required to further validate these findings.
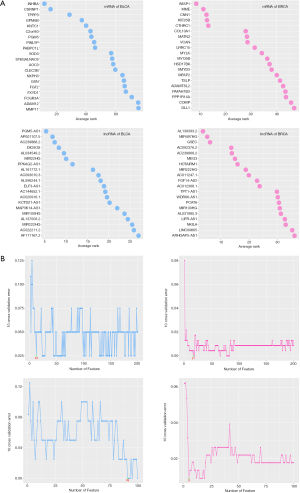
Table S1
| GeneSymbol | Degree | K-core score | Cluster | Sub-NetworkColor |
|---|---|---|---|---|
| CDK1 | 103 | 37 | Cluster1 | Blue |
| CCNB1 | 89 | 37 | Cluster1 | Blue |
| CCNB2 | 71 | 37 | Cluster1 | Blue |
| PLK1 | 69 | 37 | Cluster1 | Blue |
| CDC20 | 68 | 37 | Cluster1 | Blue |
| AURKB | 67 | 37 | Cluster1 | Blue |
| BUB1 | 65 | 37 | Cluster1 | Blue |
| MAD2L1 | 63 | 37 | Cluster1 | Blue |
| CENPE | 58 | 37 | Cluster1 | Blue |
| KIF2C | 57 | 37 | Cluster1 | Blue |
| CDCA8 | 53 | 37 | Cluster1 | Blue |
| CENPA | 52 | 37 | Cluster1 | Blue |
| BUB1B | 52 | 37 | Cluster1 | Blue |
| KIF18A | 50 | 37 | Cluster1 | Blue |
| NDC80 | 50 | 37 | Cluster1 | Blue |
| BIRC5 | 49 | 37 | Cluster1 | Blue |
| CENPF | 47 | 37 | Cluster1 | Blue |
| MLF1IP | 45 | 37 | Cluster1 | Blue |
| CENPO | 45 | 37 | Cluster1 | Blue |
| CENPN | 45 | 37 | Cluster1 | Blue |
| CENPM | 45 | 37 | Cluster1 | Blue |
| CENPL | 45 | 37 | Cluster1 | Blue |
| CENPK | 45 | 37 | Cluster1 | Blue |
| CENPI | 45 | 37 | Cluster1 | Blue |
| CASC5 | 45 | 37 | Cluster1 | Blue |
| NUF2 | 41 | 37 | Cluster1 | Blue |
| ERCC6L | 40 | 37 | Cluster1 | Blue |
| SKA1 | 39 | 37 | Cluster1 | Blue |
| ZWINT | 38 | 37 | Cluster1 | Blue |
| ZWILCH | 38 | 37 | Cluster1 | Blue |
| SPC25 | 38 | 37 | Cluster1 | Blue |
| SPC24 | 38 | 37 | Cluster1 | Blue |
| SGOL2 | 38 | 37 | Cluster1 | Blue |
| SGOL1 | 38 | 37 | Cluster1 | Blue |
| RCC2 | 38 | 37 | Cluster1 | Blue |
| KNTC1 | 38 | 37 | Cluster1 | Blue |
| DSN1 | 38 | 37 | Cluster1 | Blue |
| CDCA5 | 38 | 37 | Cluster1 | Blue |
| ESPL1 | 41 | 36 | Cluster1 | Blue |
| ADCY5 | 34 | 17 | Cluster2 | Pink |
| ADCY4 | 33 | 17 | Cluster2 | Pink |
| GNG2 | 33 | 17 | Cluster2 | Pink |
| GNG7 | 32 | 17 | Cluster2 | Pink |
| GNG11 | 32 | 17 | Cluster2 | Pink |
| LPAR2 | 26 | 17 | Cluster2 | Pink |
| C3 | 24 | 17 | Cluster2 | Pink |
| CXCL12 | 22 | 17 | Cluster2 | Pink |
| CCL21 | 18 | 17 | Cluster2 | Pink |
| SSTR1 | 18 | 17 | Cluster2 | Pink |
| GPER | 18 | 17 | Cluster2 | Pink |
| S1PR1 | 17 | 17 | Cluster2 | Pink |
| P2RY14 | 17 | 17 | Cluster2 | Pink |
| OXER1 | 17 | 17 | Cluster2 | Pink |
| CXCL2 | 17 | 17 | Cluster2 | Pink |
| ADRA2C | 17 | 17 | Cluster2 | Pink |
| ADRA2B | 17 | 17 | Cluster2 | Pink |
| ADRA2A | 17 | 17 | Cluster2 | Pink |
| IGF1 | 21 | 13 | Cluster3 | Orange |
| VWF | 18 | 13 | Cluster3 | Orange |
| A2M | 17 | 13 | Cluster3 | Orange |
| HGF | 16 | 13 | Cluster3 | Orange |
| CLU | 15 | 13 | Cluster3 | Orange |
| SERPING1 | 15 | 13 | Cluster3 | Orange |
| FIGF | 15 | 13 | Cluster3 | Orange |
| CFD | 14 | 13 | Cluster3 | Orange |
| F8 | 14 | 13 | Cluster3 | Orange |
| RARRES2 | 13 | 13 | Cluster3 | Orange |
| PROS1 | 13 | 13 | Cluster3 | Orange |
| MMRN1 | 13 | 13 | Cluster3 | Orange |
| GAS6 | 13 | 13 | Cluster3 | Orange |
| CLEC3B | 13 | 13 | Cluster3 | Orange |
| POLE2 | 26 | 12 | Cluster4 | Green |
| GEN1 | 12 | 12 | Cluster4 | Green |
| PCNA | 43 | 11.86813187 | Cluster4 | Green |
| BRCA1 | 33 | 11.86813187 | Cluster4 | Green |
| BLM | 30 | 11.86813187 | Cluster4 | Green |
| RAD51 | 23 | 11.86813187 | Cluster4 | Green |
| RMI1 | 22 | 11.86813187 | Cluster4 | Green |
| BRCA2 | 21 | 11.86813187 | Cluster4 | Green |
| EXO1 | 19 | 11.86813187 | Cluster4 | Green |
| DNA2 | 19 | 11.86813187 | Cluster4 | Green |
| RMI2 | 17 | 11.86813187 | Cluster4 | Green |
| BRIP1 | 17 | 11.86813187 | Cluster4 | Green |
| XRCC2 | 14 | 11.86813187 | Cluster4 | Green |
| RAD51AP1 | 14 | 11.86813187 | Cluster4 | Green |
| EME1 | 18 | 11 | Cluster4 | Green |
| CLSPN | 20 | 10 | Cluster4 | Green |
| SOCS3 | 16 | 10 | Cluster4 | Green |
| CCNF | 13 | 10 | Cluster4 | Green |
| KLHL13 | 13 | 10 | Cluster4 | Green |
| UBE2S | 13 | 10 | Cluster4 | Green |
| PARK2 | 11 | 10 | Cluster4 | Green |
| TRAIP | 11 | 10 | Cluster4 | Green |
| ZBTB16 | 10 | 10 | Cluster4 | Green |
| KLHL21 | 10 | 10 | Cluster4 | Green |
| ASB1 | 10 | 10 | Cluster4 | Green |
| ORC1 | 19 | 9.848484848 | Cluster4 | Green |
| MCM10 | 17 | 9.848484848 | Cluster4 | Green |
| ORC6 | 17 | 9.848484848 | Cluster4 | Green |
| DBF4 | 12 | 9.848484848 | Cluster4 | Green |
| H2AFX | 41 | 15 | Cluster5 | Yellow |
| HIST2H2BE | 39 | 15 | Cluster5 | Yellow |
| HIST1H2BD | 39 | 15 | Cluster5 | Yellow |
| STRA13 | 22 | 15 | Cluster5 | Yellow |
| OIP5 | 15 | 15 | Cluster5 | Yellow |
| MIS18A | 15 | 15 | Cluster5 | Yellow |
| HJURP | 15 | 15 | Cluster5 | Yellow |
| AURKA | 38 | 14.90333333 | Cluster5 | Yellow |
| NUSAP1 | 23 | 14.55882353 | Cluster5 | Yellow |
| DLGAP5 | 33 | 14.40692641 | Cluster5 | Yellow |
| TTK | 23 | 14.31372549 | Cluster5 | Yellow |
| KIF11 | 44 | 14.12169312 | Cluster5 | Yellow |
| KIF4A | 22 | 14 | Cluster5 | Yellow |
| KIF15 | 19 | 14 | Cluster5 | Yellow |
| KIFC2 | 14 | 14 | Cluster5 | Yellow |
| KIFC1 | 14 | 14 | Cluster5 | Yellow |
| KIF26B | 14 | 14 | Cluster5 | Yellow |
| KIF26A | 14 | 14 | Cluster5 | Yellow |
| KIF22 | 14 | 14 | Cluster5 | Yellow |
| KIF18B | 14 | 14 | Cluster5 | Yellow |
| KIF23 | 37 | 13.71428571 | Cluster5 | Yellow |
| KIF20A | 39 | 13.44 | Cluster5 | Yellow |
| CCNA2 | 55 | 13.37662338 | Cluster5 | Yellow |
| PRC1 | 29 | 13.31168831 | Cluster5 | Yellow |
| UBE2C | 36 | 13 | Cluster5 | Yellow |
| TOP2A | 44 | 12.6 | Cluster5 | Yellow |
| TPX2 | 27 | 12.45833333 | Cluster5 | Yellow |
Table S2
| Cancer | Dataset | Method | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|
| BLCA | mRNA | NB | 1 | 1 | 1 |
| SVM | 1 | 1 | 1 | ||
| K-NN | 1 | 1 | 1 | ||
| ANN | 1 | 1 | 1 | ||
| lncRNA | NB | 0.99 | 0.99 | 1 | |
| SVM | 1 | 1 | 1 | ||
| K-NN | 0.98 | 1 | 0.96 | ||
| ANN | 1 | 1 | 1 | ||
| BRCA | mRNA | NB | 1 | 1 | 1 |
| SVM | 1 | 1 | 1 | ||
| K-NN | 1 | 1 | 1 | ||
| ANN | 1 | 1 | 1 | ||
| lncRNA | NB | 1 | 1 | 1 | |
| SVM | 0.99 | 1 | 0.98 | ||
| K-NN | 0.99 | 1 | 0.98 | ||
| ANN | 0.99 | 0.98 | 1 |
BLCA, bladder cancer; BRCA, breast cancer.
Acknowledgments
Funding: This work is supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.92). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sanli O, Dobruch J, Knowles MA, et al. Bladder cancer. Nat Rev Dis Primers 2017;3:17022. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61. [Crossref] [PubMed]
- McConkey DJ, Choi W, Dinney CP. New insights into subtypes of invasive bladder cancer: considerations of the clinician. Eur Urol 2014;66:609-10. [Crossref] [PubMed]
- Aronson SJ, Rehm HL. Building the foundation for genomics in precision medicine. Nature 2015;526:336-42. [Crossref] [PubMed]
- Damrauer JS, Hoadley KA, Chism DD, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A 2014;111:3110-5. [Crossref] [PubMed]
- Cardoso F, van't Veer LJ, Bogaerts J, et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N Engl J Med 2016;375:717. [Crossref] [PubMed]
- van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002;415:530. [Crossref] [PubMed]
- Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA–LncRNA Interactions. Methods Mol Biol 2016;1402:271. [Crossref] [PubMed]
- Wang Y, Hou J, He D, et al. The Emerging Function and Mechanism of ceRNAs in Cancer. Trends Genet 2016;32:211-24. [Crossref] [PubMed]
- Nakayama S, Torikoshi Y, Takahashi T, et al. Prediction of paclitaxel sensitivity by CDK1 and CDK2 activity in human breast cancer cells. Breast Cancer Res 2009;11:R12. [Crossref] [PubMed]
- Yang F, Luo LJ, Zhang L, et al. MiR-346 promotes the biological function of breast cancer cells by targeting SRCIN1 and reduces chemosensitivity to docetaxel. Gene 2017;600:21-8. [Crossref] [PubMed]
- Jadaliha M, Zong X, Malakar P, et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget 2016;7:40418-36. [Crossref] [PubMed]
- Ford D, Easton DF, Stratton M, et al. Genetic Heterogeneity and Penetrance Analysis of the BRCA1 and BRCA2 Genes in Breast Cancer Families. Am J Hum Genet 1998;62:676-89. [Crossref] [PubMed]
- Huang M, Zhong Z, Lv M, et al. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget 2016;7:47186-200. [PubMed]
- Floyd RV, Wray S, Martín-Vasallo P, et al. Differential cellular expression of FXYD1 (phospholemman) and FXYD2 (gamma subunit of Na, K-ATPase) in normal human tissues: a study using high density human tissue microarrays. Ann Anat 2010;192:7-16. [Crossref] [PubMed]
- Font A, Taron M, Gago JL, et al. BRCA1 mRNA expression and outcome to neoadjuvant cisplatin-based chemotherapy in bladder cancer. Ann Oncol 2011;22:139-44. [Crossref] [PubMed]
- Ravasz E, Somera AL, Mongru DA, et al. Hierarchical Organization of Modularity in Metabolic Networks. Science 2002;297:1551-5. [Crossref] [PubMed]
- Ying L, Huang Y, Chen H, et al. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst 2013;9:407-11. [Crossref] [PubMed]
- Breiman L. Random Forests. Machine Learning 2001;45:5-32. [Crossref]
- Li R, Qu H, Wang S, et al. GDCRNATools: an R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics 2018;34:2515-7. [Crossref] [PubMed]
- Okegawa T, Li Y, Pong RC, et al. Cell adhesion proteins as tumor suppressors. J Urol 2002;167:1836-43. [Crossref] [PubMed]
- Syrigos KN, Harrington KJ, Pignatelli M. Role of adhesion molecules in bladder cancer: an important part of the jigsaw. Urology 1999;53:428-34. [Crossref] [PubMed]
- Bergamaschi A, Tagliabue E, Sørlie T, et al. Extracellular matrix signature identifies breast cancer subgroups with different clinical outcome. J Pathol 2008;214:357-67. [Crossref] [PubMed]
- Gordon JN, Shu WP, Schlussel RN, et al. Altered extracellular matrices influence cellular processes and nuclear matrix organizations of overlying human bladder urothelial cells. Cancer Res 1993;53:4971-7. [PubMed]
- Lochter A, Bissell MJ. Involvement of extracellular matrix constituents in breast cancer. Semin Cancer Biol 1995;6:165-73. [Crossref] [PubMed]
- Luo M, Guan JL. Focal adhesion kinase: a prominent determinant in breast cancer initiation, progression and metastasis. Cancer Lett 2010;289:127-39. [Crossref] [PubMed]
- Kong DB, Chen F, Sima N. Focal adhesion kinases crucially regulate TGFβ-induced migration and invasion of bladder cancer cells via Src kinase and E-cadherin. Onco Targets Ther 2017;10:1783-92. [Crossref] [PubMed]
- Golubovskaya VM, Ylagan L, Miller A, et al. High focal adhesion kinase expression in breast carcinoma is associated with lymphovascular invasion and triple-negative phenotype. BMC Cancer 2014;14:769. [Crossref] [PubMed]
- Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol 2001;2:21-32. [Crossref] [PubMed]
- Song Y, Zhao C, Dong L, et al. Overexpression of cyclin B1 in human esophageal squamous cell carcinoma cells induces tumor cell invasive growth and metastasis. Carcinogenesis 2008;29:307. [Crossref] [PubMed]
- Choi JW, Kim Y, Lee JH, et al. High expression of spindle assembly checkpoint proteins CDC20 and MAD2 is associated with poor prognosis in urothelial bladder cancer. Virchows Arch 2013;463:681-7. [Crossref] [PubMed]
- Karra H, Repo H, Ahonen I, et al. Cdc20 and securin overexpression predict short-term breast cancer survival. Br J Cancer 2014;110:2905-13. [Crossref] [PubMed]
- Yamamoto Y, Matsuyama H, Chochi Y, et al. Overexpression of BUBR1 is associated with chromosomal instability in bladder cancer. Cancer Genet Cytogenet 2007;174:42-7. [Crossref] [PubMed]
- Han JY, Han YK, Park GY, et al. Bub1 is required for maintaining cancer stem cells in breast cancer cell lines. Sci Rep 2015;5:15993. [Crossref] [PubMed]
- Sun L, Li Y, Yang B. Downregulated long non-coding RNA MEG3 in breast cancer regulates proliferation, migration and invasion by depending on p53's transcriptional activity. Biochem Biophys Res Commun 2016;478:323-9. [Crossref] [PubMed]
- Ning X, Deng Y. Identification of key pathways and genes influencing prognosis in bladder urothelial carcinoma. Onco Targets Ther 2017;10:1673-86. [Crossref] [PubMed]
- Lawrence RT, Perez EM, Miller CP, et al. The Proteomic Landscape of Triple-Negative Breast Cancer. Cell Rep 2015;11:990. [Crossref] [PubMed]
- Rad A, Farshchian M, Forghanifard MM, et al. Predicting the molecular role of MEIS1 in esophageal squamous cell carcinoma. Tumour Biol 2016;37:1715-25. [Crossref] [PubMed]
- Beukers W, Hercegovac A, Vermeij M, et al. Hypermethylation of the polycomb group target gene PCDH7 in bladder tumors from patients of all ages. J Urol 2013;190:311-6. [Crossref] [PubMed]
- Doolan P, Clynes M, Kennedy S, et al. TMEM25, REPS2 and Meis 1: favourable prognostic and predictive biomarkers for breast cancer. Tumour Biol 2009;30:200-9. [Crossref] [PubMed]
- Schäfer MKE, Altevogt P. L1CAM malfunction in the nervous system and human carcinomas. Cell Mol Life Sci 2010;67:2425-37. [Crossref] [PubMed]
- Faltas BM, Prandi D, Tagawa ST, et al. Clonal Evolution of Chemotherapy-resistant Urothelial Carcinoma. Nature Genet 2016;48:1490. [Crossref] [PubMed]
- Doberstein K, Milde-Langosch K, Bretz NP, et al. L1CAM is expressed in triple-negative breast cancers and is inversely correlated with androgen receptor. BMC Cancer 2014;14:958. [Crossref] [PubMed]
- Han HJ, Russo J, Kohwi Y, et al. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature 2008;452:187. [Crossref] [PubMed]
- Wan F, Cheng C, Wang Z, et al. SATB1 overexpression regulates the development and progression in bladder cancer through EMT. Plos One 2015;10:e0117518. [Crossref] [PubMed]
- Patani N, Jiang W, Mansel R, et al. The mRNA expression of SATB1 and SATB2 in human breast cancer. Cancer Cell International 2009;9:18. [Crossref] [PubMed]
- Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: from function to translation. Trends Cancer 2015;1:93-109. [Crossref] [PubMed]
- Choi W, Porten S, Kim S, et al. Identification of Distinct Basal and Luminal Subtypes of Muscle-Invasive Bladder Cancer with Different Sensitivities to Frontline Chemotherapy. Cancer Cell 2014;25:152-65. [Crossref] [PubMed]
- Blaustein MP. Calcium as an Intracellular Second Messenger. In: Rubin RP, Weiss GB, Putney JW Jr. Calcium in Biological Systems. Elsevier Inc, 2001.
- Pedrocchi M, Schäfer BW, Mueller H, et al. Expression of Ca2+‐binding proteins of the S100 family in malignant human breast‐cancer cell lines and biopsy samples. Int J Cancer 1994;57:684-90. [Crossref] [PubMed]
- Jiang H, Hu H, Tong X, et al. Calcium-binding protein S100P and cancer: mechanisms and clinical relevance. J Cancer Res Clin Oncol 2012;138:1-9. [Crossref] [PubMed]
- Parker JS, Mullins M, Cheang MCU, et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J Clin Oncol 2009;27:1160. [Crossref] [PubMed]
- Duan KB, Rajapakse JC, Wang H, et al. Multiple SVM-RFE for gene selection in cancer classification with expression data. IEEE Transactions on Nanobioscience 2005;4:228. [Crossref] [PubMed]
- Zhang M, Wang W, Li T, et al. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother 2016;80:73-9. [Crossref] [PubMed]
- Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nature Reviews Cancer 2009;9:785. [Crossref] [PubMed]
- Tang Y, Simoneau AR, Liao WX, et al. WIF1, a Wnt pathway inhibitor, regulates SKP2 and c-myc expression leading to G1 arrest and growth inhibition of human invasive urinary bladder cancer cells. Mol Cancer Ther 2009;8:458. [Crossref] [PubMed]
- Traub F, Mengel M, Lück HJ, et al. Prognostic impact of Skp2 and p27 in human breast cancer. Breast Cancer Res Treat 2006;99:185-91. [Crossref] [PubMed]
- Zhang Y, Cheng S, Zhang G, et al. Low expression of RECK indicates a shorter survival for patients with invasive breast cancer. Cancer Sci 2012;103:1084-9. [Crossref] [PubMed]
- Stavik B, Skretting G, Aasheim HC, et al. Downregulation of TFPI in breast cancer cells induces tyrosine phosphorylation signaling and increases metastatic growth by stimulating cell motility. BMC Cancer 2011;11:357. [Crossref] [PubMed]
- Umetani N, Mori T, Koyanagi K, et al. Aberrant hypermethylation of ID4 gene promoter region increases risk of lymph node metastasis in T1 breast cancer. Oncogene 2005;24:4721-7. [Crossref] [PubMed]


