
CXCL8 is a potential biomarker for predicting disease progression in gastric carcinoma
Introduction
Gastric cancer (GC) is one of the leading causes of cancer-related mortality worldwide (1), and has the second highest incidence rate of all cancers (2,3). As the most common malignancy in the digestive system, GC is a major public health concern (4), especially in China which is among the countries with the highest incidence of GC (5-7). Although the etiology of GC has not been fully elucidated, its pathogenesis is believed to be associated with extremely complicated processes that involve multiple factors (8). To a large extent, early diagnosis significantly determines the prognosis of GC, as the 5-year survival rate of early GC is more than 90%, while that of advanced GC is less than 50% (9). Therefore, timely detection of GC-specific tumor markers can assist early diagnosis, reduce mortality rate, and improve prognosis of GC patients. However, because of the lack of specific symptoms and signs in early stages of GC, it is difficult to diagnose most patients. Exploring economic, effective, and safe screening methods for the early diagnosis of GC and predicting the prognosis of GC patients is extremely important.
CXC chemokine 8 (CXCL8), which belongs to the family of CXC chemokines (C is cysteine, X refers to an arbitrary amino acid), is generated by many cells (10). CXCL8 has a chemotactic effect on specific white blood cells and can be stimulated by phytohemagglutinin, lipopolysaccharide, bacterial virus products, tumor necrosis factor-α (TNF-α), and interleukin (IL). Also known as IL-8, CXCL8 activates and promotes a series of inflammatory responses, such as chemotaxis and degranulation of neutrophils (11). CXCL8 is abnormally expressed in many tumors and is involved in tumor development (12). In addition to chemotaxis and activation of immune cells, CXCL8 expression is closely related to the occurrence, development, and metastasis of cancer cells in many tumors (13). However, the diagnostic and prognostic values of CXCL8 in GC have not been extensively studied.
In the current study, we explored potential prognostic predictors and therapeutic targets by systematically analyzing the gene expression profiles of GC and control samples using the Cancer Genome Atlas (TCGA) database, and found that CXCL8 is up-regulated in GC. We further validated the CXCL8 expression pattern in the Oncomine database, a cancer microarray database, and a web-based data-mining platform (14). Our subsequent survival analysis suggests that CXCL8 is a potential biomarker for predicting progress of GC.
Methods
Databases
Data for the GC tissue (375 cases, GC group) and neighboring noncancerous adrenal tissues (32 cases, Control group) with complete clinical and survival information were downloaded from the TCGA database (https://cancergenome.nih.gov/). The RStudio-1.1.419 software was employed to analyze the differential expression levels between the two groups. The Oncomine database (https://www.oncomine.org) (14) was also selected to search the differential expression levels of target genes between groups. Additionally, the Kaplan-Meier Plotter database (http://kmplot.com/analysis/index.php?p=service&cancer=gastric) (15) was used to generate the overall survival (OS) and disease-free survival (DFS) curves.
Functional and pathway enrichment analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology Based Annotation System (https://david.ncifcrf.gov/), based on multiple databases including pathways, diseases, and gene ontology (GO), was used to execute KEGG pathway enrichment analysis and GO enrichment analysis (16,17).
Protein-protein interaction (PPI) network construction
The RStudio Tool was used to generate PPI data (18). In addition, the database was employed to quantify the interrelationships of analyzed differentially expressed genes (DEGs). Results were visualized using Cytoscape software (19). A confidence score >0.4 was defined as significant. The hub genes were those that had a connectivity degree over 20 in the PPI network.
Prognostic value of each hub gene
Based on the TCGA database, the RStudio-1.1.419 software was used to investigate the impacts of the expression levels of each hub gene on OS or DFS of GC patients. The log-rank test and the Mantel-Cox test were used as hypothesis tests. The cohort thresholds were adjusted and gene-pairs were used.
Survival analyses
The Kaplan plot univariate and Cox multivariate survival analyses were used to verify the correlation between gene expression and clinicopathological parameters in GC patients, as well as their clinical prognostic significance. The survival plots also contained the cox proportional hazard ratio and P=0.05 was set as the significance level.
Immunohistochemistry (IHC) assay
The IHC assay data from GC and normal gastric tissues were downloaded from The Human Protein Atlas (https://www.proteinatlas.org/ENSG00000169429-CXCL8/tissue/stomach). A polyclonal antibody against human CXCL8 (HPA057179) was used for the IHC assay.
Statistical analysis
SPSS 22.0 software (IBM, Armonk, NY, USA) was used for statistical analysis. The Chi-square test and Fisher exact probability method were used to analyze the correlations between the expression levels of biomarkers and the clinicopathological characteristics of patients. Statistical significance was defined as P<0.05.
Results
Identification of DEGs between GC and healthy control samples
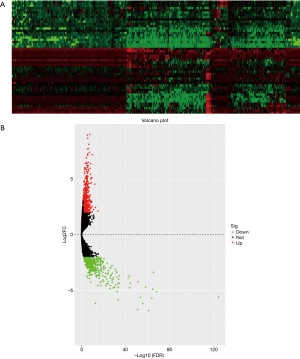
First, we performed gene expression profiling for 375 GC tissues and 32 normal gastric tissues, whose information was deposited in the TCGA database. After quality assessment and data preprocessing, such as a rank aggregation analysis of the expression matrices, we identified 1,313 statistically significantly altered genes, including 781 up-regulated and 532 down-regulated genes, in GC samples compared to control tissues. The heatmap (Figure 1A) and Volcano plot (Figure 1B) show the up-regulated DEGs (red) and down-regulated DEGs (green).
Functional and pathway DEG enrichment analysis
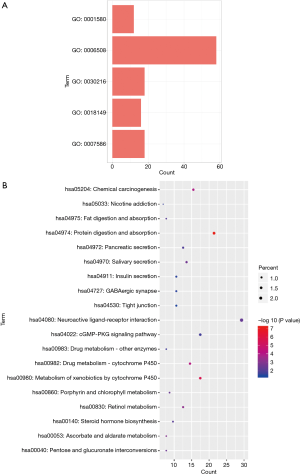
To gain further insight about the function of the identified DEGs, all DEGs were subjected to pathway enrichment analysis (https://david.ncifcrf.gov/). The GO analysis indicated that five biological processes were enriched: Go: 0001580, Go: 0006508, Go: 0030216, Go: 0018149, and Go: 0007586 (all P value <0.05) (Figure 2A). Furthermore, 19 significantly enriched KEGG pathways (hsa05204, hsa05033, hsa04975, hsa04974, hsa04972, hsa04970, hsa04911, hsa04727, hsa04530, sha04080, hsa04002, sha00983, hsa00982, hsa00980, hsa00860, hsa00830, hsa00140, hsa000530, and hsa00040) were identified (all P value <0.05). The top 5 most significantly enriched KEGG pathways were as follows: hsa04972, protein digestion and absorption; hsa00980, metabolism of xenobiotics by cytochrome P450; hsa05204, chemical carcinogenesis; hsa00982, drug metabolism-cytochrome P450; and hsa00830, retinol metabolism (Figure 2B).
Identification of hub genes using PPI network analysis of DEGs
We constructed a PPI network using a cutoff confidence >0.4 and degree of connectivity >20. We found 20 genes that were regarded as hub genes among the 1,313 DEGs (Figure 3). The top 10 hub genes—GNGT1, KNG1, GNG7, ANXA1, GPR17, LPAR3, PPBP, CXCL8, NPY, and SST—were chosen for further analysis.
Assessment of hub genes in the TCGA and Oncomine database
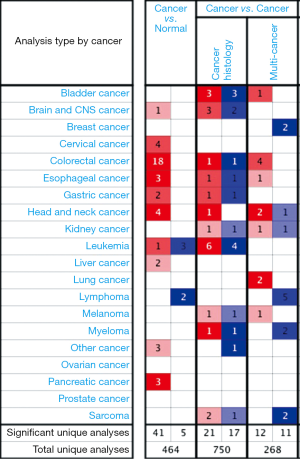
To assess the roles of the ten hub genes in GC, we validated the gene expression using both the TCGA and Oncomine databases. CXCL8 was the only gene that was up-regulated in tumor tissues in both the TCGA and Oncomine databases. In the comparison analysis, the mRNA levels of CXCL8 were also up-regulated in multiple cancer types (Figure 4) and in cancer tissues from different cohorts of patients with GC (Figure 5) compared to controls. In addition, we analyzed genes that potentially correlated with CXCL8 in the GC data in the Oncomine database. Twenty genes—AREG, HBEGF, IL6, IL1B, PTGS2, KLF6, FOS, FOSB, MAP3KB, ETS2, FOSL2, LIF, PIM1, OSM, IL1RN, CSF3, WEE1, CTGF, SERPINE1, and MLF1—correlated with CXCL8 in GC (Figure 6).
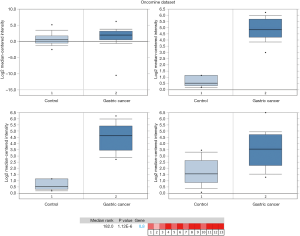

Higher expression of CXCL8 in GC tissues predicts worse survival
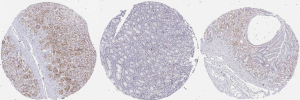
We further validated CXCL8 protein expression in clinical samples from the tumor tissues of GC patients. As shown in Figure 7, IHC analysis suggested medium expression of both cytoplasmic and membranous CXCL8 in GC tissues. To reveal the clinical prognostic significance of CXCL8 expression, we used the Kaplan plot univariate and Cox multivariate survival analyses to verify the correlation between CXCL8 mRNA levels and clinicopathological parameters in GC patients. The patients with lower CXCL8 mRNA levels in the tumor tissues displayed significantly (P<0.001) prolonged relapse-free survival (RFS) compared to the patients with higher CXCL8 mRNA levels in tumor tissues (Figure 8). Therefore, CXCL8 is a potential biomarker for predicting disease progress and prognosis in GC.

Discussion
There are more than ten immunohistochemical markers that can be used to clinically diagnose GC and predict the prognosis of GC patients. However, markers with high specificity and sensitivity are still lacking (20). In this study, we conducted integrated bioinformatics analysis of gene expression datasets in GC and control samples using the TCGA and Oncomine databases, and screened hub genes that might be involved in GC pathogenesis. We found that CXCL8 mRNA levels were increased in GC tissues compared to normal gastric tissues, and that GC patients with higher CXCL8 tumor expression had significantly shortened RFS. Thus, we conclude that CXCL8 is a potential novel biomarker that could be used for the early diagnosis of GC and prognosis prediction of patients with GC.
CXCL8 can contribute to the tumorigenesis and pathogenesis of GC in many aspects. CXCL8 is predominantly secreted by macrophages and contributes to the immunosuppressive microenvironment by inducing PD-L1+ macrophages in GC (10). Consistent with our conclusion, CXCL8 inhibitors have been reported to promote an anti-tumor response, providing potential therapeutic effects for patients with GC (10). In addition, CXCL8 derived from tumors can activate endothelial cells in tumor vessels, promote angiogenesis, and induce neutrophil chemotaxis to allow immune cell infiltration into the tumor site (21,22). Therefore, CXCL8 can both promote the invasion and migration of cancer cells and induce macrophages to secrete additional growth factors, both of which further increase the proliferation and invasion rate of cancer cells. Therefore, inhibition of CXCL8 in the tumor microenvironment has significant potential for tumor treatment (23).
CXCL8 is a potential biomarker for predicting tumor progression in other malignancies. For example, CXCL8 is continuously expressed in melanoma and is up-regulated in melanoma cells with high metastasis potential (24). In breast cancer, CXCL8 has been identified as a key factor involved in metastasis (25). In human ovarian cancer cell lines with high metastasis, CXCL8 expression was higher than that of parental cell lines (26). One study demonstrated that the proliferation rate of ovarian cancer epithelial cell lines with high expression of CXCL8 was significantly faster compared to cell lines with low CXCL8 expression (26). Furthermore, the expression level of CXCL8 in the PC-3M-LN4 cell line with high metastasis capability was significantly higher compared to the PC-3P cell line with low metastasis capability. Overexpression of CXCL8 could significantly promote the tumorigenicity, metastasis, and angiogenesis of PC-3P cells in nude mice (27). Taken together, these studies indicate a pervasive role of CXCL8 in promoting tumorigenesis, metastasis, and tumor survival.
This study has some limitations. First, the TCGA and Oncomine databases provide data at the RNA level, which may not accurately reflect protein expression demonstrated by traditionally pathological IHC. Therefore, the bioinformatics data shown in this study cannot fully represent the actual protein expression of CXCL8. The clinical value of CXCL8 in GC should be analyzed using IHC in future studies. Secondly, this study mainly excavates the related data and analysis based on the information provided by online databases; thus, our conclusion needs to be further validated using relevant, actual clinical research results. Despite these limitations, we believe our work provides complementary evidence on the potential use of CXCL8 for diagnosis and prognosis prediction of GC patients, as well as targeting CXCL8 for GC therapy.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.12.52). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ren JS, Masuyer E, et al. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer 2013;132:1133-45. [Crossref] [PubMed]
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Zong L, Abe M, Seto Y, et al. The challenge of screening for early gastric cancer in China. Lancet 2016;388:2606. [Crossref] [PubMed]
- Strong VE, Wu AW, Selby LV, et al. Differences in gastric cancer survival between the U.S. and China. J Surg Oncol 2015;112:31-7. [Crossref] [PubMed]
- Kim YI, Choi IJ, Kook MC, et al. The association between Helicobacter pylori status and incidence of metachronous gastric cancer after endoscopic resection of early gastric cancer. Helicobacter 2014;19:194-201. [Crossref] [PubMed]
- Karimi P, Islami F, Anandasabapathy S, et al. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev 2014;23:700-13. [Crossref] [PubMed]
- Li Z, Zhao Y, Liu Y, et al. Laparoscopic versus open gastrectomy for high-risk patients with gastric cancer: A systematic review and meta-analysis. Int J Surg 2019;65:52-60. [Crossref] [PubMed]
- Lin C, He H, Liu H, et al. Tumour-associated macrophages-derived CXCL8 determines immune evasion through autonomous PD-L1 expression in gastric cancer. Gut 2019;68:1764-73. [Crossref] [PubMed]
- Ruffini PA. The CXCL8-CXCR1/2 Axis as a Therapeutic Target in Breast Cancer Stem-Like Cells. Front Oncol 2019;9:40. [Crossref] [PubMed]
- Fisher RC, Bellamkonda K, Alex Molina L, et al. Disrupting Inflammation-Associated CXCL8-CXCR1 Signaling Inhibits Tumorigenicity Initiated by Sporadic- and Colitis-Colon Cancer Stem Cells. Neoplasia 2019;21:269-81. [Crossref] [PubMed]
- Ha H, Debnath B, Neamati N. Role of the CXCL8-CXCR1/2 Axis in Cancer and Inflammatory Diseases. Theranostics 2017;7:1543-88. [Crossref] [PubMed]
- Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. [Crossref] [PubMed]
- Szasz AM, Lanczky A, Nagy A, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 2016;7:49322-33. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Panarese I, De Vita F, Ronchi A, et al. Predictive biomarkers along gastric cancer pathogenetic pathways. Expert Rev Anticancer Ther 2017;17:417-25. [Crossref] [PubMed]
- Dhanani AS, Bagchi T. Lactobacillus plantarum CS24.2 prevents Escherichia coli adhesion to HT-29 cells and also down-regulates enteropathogen-induced tumor necrosis factor-alpha and interleukin-8 expression. Microbiol Immunol 2013;57:309-15. [Crossref] [PubMed]
- Mao W, Peters HL, Sutton MN, et al. The role of vascular endothelial growth factor, interleukin 8, and insulinlike growth factor in sustaining autophagic DIRAS3-induced dormant ovarian cancer xenografts. Cancer 2019;125:1267-80. [Crossref] [PubMed]
- Crespo J, Wu K, Li W, et al. Human Naive T Cells Express Functional CXCL8 and Promote Tumorigenesis. J Immunol 2018;201:814-20. [Crossref] [PubMed]
- Wu S, Singh S, Varney ML, et al. Modulation of CXCL-8 expression in human melanoma cells regulates tumor growth, angiogenesis, invasion, and metastasis. Cancer Med 2012;1:306-17. [Crossref] [PubMed]
- Fang QI, Wang X, Luo G, et al. Increased CXCL8 Expression Is Negatively Correlated with the Overall Survival of Patients with ER-Negative Breast Cancer. Anticancer Res 2017;37:4845-52. [PubMed]
- Milliken D, Scotton C, Raju S, et al. Analysis of chemokines and chemokine receptor expression in ovarian cancer ascites. Clin Cancer Res 2002;8:1108-14. [PubMed]
- Inoue K, Slaton JW, Eve BY, et al. Interleukin 8 expression regulates tumorigenicity and metastases in androgen-independent prostate cancer. Clin Cancer Res 2000;6:2104-19. [PubMed]


