
Strategies for enrichment of circulating tumor cells
Introduction
It has generally been believed that the dissemination of tumor to various organs occurs owing to tumor cells dissociating from the primary sites into the peripheral blood and then settling and growing at distant sites, finally resulting in tumor recurrence. Circulating tumor cells (CTCs) was first verified by Ashworth who microscopically observed cells resembled tumor cells in 1869 in a metastatic patient’s peripheral blood (1). In 1954, CTCs was shown to have significant metastatic potential (2). As a form of ‘liquid biopsy’, CTCs are noninvasive and safe compared with traditional tissue biopsy, which can be used for real-time monitoring of therapy response and tumor progression. Some studies have shown that CTCs can be detected (3,4), while the primary tumor mass is not observed by conventional imaging methods owing to their limitation of sensitivity during the early stages of tumor development (5). The demand for clinical utility of CTCs, e.g., early diagnosis, prediction of prognosis, assessment of recurrent risk, supervision of curative effects and individualized treatment has pushed forward research in this field. However, studies of CTCs have encountered obstacles for years due to their extremely low concentrations, with occurrence of 1–10 cells per 10-mL of blood (far fewer than billions of hematopoietic cells) (6) and short half-life (approximately from 1 to 2.4 h) in blood (7), which make subsequent analysis difficult. Moreover, only a limited number of CTCs have metastatic capacity (8). Therefore, it is essential to characterize them exactly to be able to differentiate metastatic CTCs from the nonmetastatic ones. Various techniques have been developed for the effectively isolation and identification of CTCs. These methods are mostly based on the physical properties and the diverse molecular biomarker profile of CTCs. Although several methods have shown the efficient capture of CTCs, blood samples require pretreatment with fixative agents in many cases, thus disabling the isolation of living CTCs for culture and functional characterization.
In this article, we reviewed the various techniques that have been developed for the capture and characterization of CTCs.
CTCs enrichment technologies
CTCs are distributed in billions of erythrocytes and leukocytes in the peripheral blood of tumor patients (1). The crucial technical difficulties are how to effectively isolate the extremely rare CTCs for subsequent precise detection and in-depth analysis. Therefore, sensitive and reliable methods of capturing CTCs are essential to CTC study. Generally speaking, CTC enrichment can be positively or negatively performed based on their physical or biological properties via positive identification or negative elimination selection methods. Here we outline the technologies which have made important progress in this field.
Label-free enrichment of CTCs
The CTC enrichment based on their physical properties is generally label-free, which is independent of the expression biomarkers represented by CTCs. In other words, CTCs are enriched by using density, size, deformability, and electric charge as selection criteria. Such enrichment methods are usually low-cost and quick (9).
Density-based centrifugation isolation
This method was as first exhibited in 1959 by Seal using centrifugation for cell separation. A whole blood sample contains multiple types of cells, e.g., numerous red blood cells (RBCs), nucleated white blood cells (WBCs: eosinophils, basophils, neutrophils, lymphocytes, and monocytes), and heterogeneous CTCs populations with variable numbers across a great many of cancers. Dr. Seal found that different cell types in whole blood exhibited the discrepancy in cell density and utilized silicone oil blends to obtain the optimal cell isolation medium (10). The whole blood was separated by centrifugation into the following layers (from bottom to top): erythrocytes, granulocytes, density gradient, buffy coat [the fraction containing mononuclear cells (MNCs), including CTCs], and plasma (Figure 1A). To date, several commercially available separation media are Ficoll-Paque (GE Healthcare, Chicago, IL, USA), Percoll (GE Healthcare), Lymphoprep (STEMCELL Technologies, Vancouver, Canada), and OncoQuick (Greiner Bio-One, Kremsmunster, Austria). Ficoll-Paque, which are mixtures of high molecular weight sucrose polymers and sodium diatrizoate, can be used for isolating peripheral blood MNCs (11). However, due to the cytotoxicity of Ficoll, the formation of cell aggregates may lead to the loss of tumor cells that migrate to the bottom of the medium (12) (Table 1). Another density medium is Percoll, which is composed of a colloidal silica particle suspension and shows decreased cytotoxicity and a wider density gradient range over Ficoll (12). In comparison with Ficoll-Paque, OncoQuick places a permeable apparatus on the separation media in a 50-mL tube to collect CTCs while permitting the erythrocytes and leukocytes to pass through depending on their different buoyancy densities during centrifugation. The mean tumor cell recovery rates for both OncoQuick and Ficoll-Paque were similar, between 70% and 90%. However, OncoQuick extremely cut down the number of captured MNCs by 632-fold (9.5×104 MNCs on average), which is much fewer than that of Ficoll-Paque (a depletion factor of 3.8 and an average number of 1.6×107 MNCs) (13,14). This brought great convenience for subsequent immunocytochemical detection. Such separation enables maintaining cell viability for subsequent culture without extra processing. A primary limitation of this type of enrichment technique is that it cannot exactly get together all the plasma after centrifugation, which results in the loss of potential CTCs. Moreover, it is likely that CTCs may move into the plasma fraction and the aggregates of CTCs fall to the bottom of the gradient (15) (Table 1). Szczerba et al. found that CTCs were associated with neutrophils and compared the transcriptome profiles of CTC-neutrophil clusters against those of CTCs alone. A number of genes that were involved in cell cycle progression and resulted in enhanced metastasis potential were detected (16). On the other hand, very small CTCs may have similar or much larger density than RBCs and be lost with low-density separation media. A previous study demonstrated that these small cells had an aggressive potential (17). In addition, this method exhibits very low purity of CTCs owing to the inability to discriminate leukocytes from CTCs (Table 1). Williams et al. have successfully built a patient-derived xenograft (PDX) mouse model by utilizing CTCs enriched from blood samples of prostate cancer (PCa) patients through density gradient centrifugation in addition to anti-CD45 antibody-labelled magnetic beads (18). This kind of model could be an available tool for further analysis of CTCs in functional studies or molecular characterization in vivo.
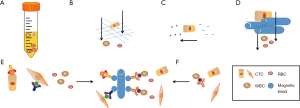
Table 1
| Technology | Selection criteria | Assay system | Advantages | Disadvantages |
|---|---|---|---|---|
| Physical property-based assays | ||||
| Density gradient centrifugation | Density | Ficoll-Paque; Percoll; Lymphoprep; OncoQuick | High cell viability; inexpensive | Loss of very small CTCs and cell aggregates; low purity extra enrichment technologies required |
| Size-based filtration exclusion | Size | ISET; DEF | Quick; non-selective | Inability to distinguish monocytes from CTCs; loss of the CTCs with similar size to WBCs |
| Deformability-based enrichment of CTCs | Deform-ability | Celsee | Quick; high sensitivity | Loss of relatively small CTCs |
| Electric-charge-based electrophoresis | DEP | ApoStream | High cell viability; high efficiency | Low purity in some devices |
| Microfluidic separation based on physical properties of CTCs | Size; deform-ability | Parsortix; ClearCell® FX1 | Short processing time of sample | The difficulty of removing the leukocytes of similar size to CTCs; the chip is easy to be blocked |
| Biological property-based assays | ||||
| Immunoaffinity-positive | EpCAM (magnetic beads) | CellSearch; MagSweeper; AdnaTest; Magwire; GILUPI; CellCollector | High recovery; high purity rates | Loss of EpCAM-negative CTCs; problems with the antibody affinity or specificity |
| EpCAM (microfluidic chips) | IsoFlux; CTC-chip; HB-chip; CMx; NanaVelcro | High purity rates; high capture efficiency; high cell viability | Long, time-consuming process; sample preprocessing requirement to reduce volume | |
| Immunoaffinity-negative | CD45; CD66b | RosetteSep; SE-iFISH; MINDEC; CTC-iChip | High cell viability; avoid loss of EpCAM -negative CTCs | Inability to deplete CD45-negative endothelial cells; loss of CTC aggregates surrounded by WBCs |
CTC, circulating tumor cell; ISET, isolation by the size of epithelial tumor; DEF, Dean flow fractionation; WBC, white blood cell; DEP, dielectrophoresis; EpCAM, epithelial cell adhesion molecule; MINDEC, multi-marker immune-magnetic negative depletion enrichment of CTCs; SE-iFISH, subtraction enrichment and immunostaining-fluorescence in-situ hybridization.
Size-based filtration exclusion
The sizes between hematological and non-hematological cells are different. Monocytes are generally larger (15–30 µm in size) than RBCs (6–8 µm) and WBCs (10–15 µm). Tumor cells possess an average size of 20 µm, higher nuclear to cytoplasm ratio and stiffness. Therefore, they are trapped by size-based filtration systems and microfluidic systems, while blood cells are not (Figure 1B). The fundamental approaches of size-based enrichment of CTCs contain isolation by the size of epithelial tumor (ISET) cells (19) and microelectromechanical system (MEMS)-based micro-filter means (20). ISET is one of the earliest size-based methods to capture CTCs by directly filtering blood through a calibrated, polycarbonate Track-Etch-type membrane with 8-µm-diameter pores. The ISET method can detect CTCs in 80 of 106 patients (75%) with advanced-stage III/IV lung adenocarcinoma and provide unmodified CTCs for determining MET status (21). Hou et al. introduced an inertial microfluidics-based separation technique for CTCs isolation, termed Dean flow fractionation (DEF), which is a spiral CTC enrichment chip that separates cells based on size; smaller blood cells migrate along Dean vortices towards the inner wall and then back to the outer wall again, whereas larger CTCs experience additional strong inertial lift forces and collect along the microchannel inner wall (11). This kind of method is characterized by quick, non-selective and label-free separation for CTCs. However, one pitfall of this technique is its inability to distinguish monocytes from CTCs in blood samples (Table 1). Furthermore, this perception is based to a great extent on the measurement of the size of cultured cell lines instead of the size of actual CTCs in human peripheral blood. The data from The National Cancer Institute (NCI) 60 human tumor cell line anticancer drug discovery project showed that the average diameter of tumor cells is (15.6±2.4) µm, while WBCs have an interval of diameters of 7–15 µm (22). The defect, however, of size-based enrichment technique of CTCs is that the sizes of many CTCs and WBCs from tumor patients resembles each other (Table 1). Actually, small CTCs have been indicative of worse disease status (17). Strategies for PCa CTC capture that are based on size exclusion result in the loss of 20–50% of CTCs (23). To eliminate this barrier, Kim et al. developed an alternative approach relying on the selective size amplification (SSA) of CTCs while utilizing a multi-obstacle architecture (MOA) filter to improve both recovery rate and purity (24). The SSA was carried out by labeling CTCs with anti-epithelial cell adhesion molecule (anti-EpCAM) antibody-coupled 3 µm magnetic beads to factitiously increase CTC diameter, leading to a much higher recovery and purity compared to natural size-based isolation.
Deformability-based enrichment of CTCs
CTCs are also enriched based on another physical property, deformability. Previous researches have shown that tumor cells are generally more deformable than hematopoietic cells (25-27). A study by Shaw Bagnall et al. compared the deformability of CTCs and hematopoietic cells (28). They evaluated the deformability by measuring the time required for the two cell types to go through a microfluidic device. Their results showed that the discrepancies in deformability between tumor cells and WBCs are greater than that between tumor cells of different metastatic potential. The above data demonstrate that diverse deformability could be used as indicators to differentiate tumor cells from WBCs. However, in a subgroup of metastatic PCa patients of the same study, CTCs have a greater mechanical similarity to blood cells than common tumor cell lines. Celsee Diagnostics has developed a technique that combines size exclusion and deformability to isolate and characterize CTCs (29,30). This device includes a parallel network of fluidic channels with 56,320 capture chambers. Larger tumor cells are retained and captured in the chambers, while smaller blood cells, e.g., RBCs and most WBCs, evades. A flaw of this method is the loss of relatively small CTCs (Table 1). CTCs could be rapidly captured in the microchannel device and used for subsequent characterization by reverse transcription-polymerase chain reaction (RT-PCR), immunocytochemistry as well as DNA or RNA fluorescence in-situ hybridization (FISH). This device facilitates label-free capture of cells, so we can use various antibodies to make further in-depth analysis of cells. Compared to the CellSearch system, CTC numbers with the Celsee system were considerably higher, indicating the latter has a greater sensitivity for CTC enrichment (30).
Electric-charge-based electrophoresis
CTCs can also be separated by dielectrophoresis (DEP) as CTCs exhibited a distinctive dielectric property. Enrichment based on differences in surface charge and polarizability minimize the injury of captured CTCs, which is favorable to future analysis. This strategy supports the assumption that tumor cells carry more negative surface charges, or higher zeta-potential, compared to WBCs (Figure 1C). A disadvantage of this method is that there is a partial overlap in the zeta-potential distribution between WBCs and CTCs, resulting in WBCs to remain in the enriched CTCs (Table 1). The ApoStream device (ApoCell, Houston, TX, USA) is a typical example, which performs the capture of CTCs relying on surface charge (31). Poklepovic et al. showed that in patients with metastatic PCa a greater account of CTCs could be isolated with this device, compared to the CellSearch test (32).
Acoustic separation of CTCs
Acoustic-based cell separation is a comprehensive label-free enrichment method that can separate cells based on their size, density, compressibility, or a combination thereof. Li et al. (33) developed an acoustic-based microfluidic device that could realize high-throughput separation of CTCs from blood samples of tumor patients by more than 20 times in comparison with previous devices. The improved method utilizes tilted-angle standing surface acoustic waves (taSSAWs) and optimizes the design parameters, such as the tilt angle and the length of the interdigitated transducers (IDTs) as well as the device power. As a result, low concentrations (~100 cells/mL) of multiple tumor cells were successfully separating from WBCs with a recovery rate of over 83%.
Microfluidic separation based on physical properties of CTCs
In recent years, it is possible to manipulate very tiny volumes of fluids in micro-and nano-scale space with the development of microfluidic technology. Microfluidic chip is the main platform and device of microfluidic technology. Owing to its small size, controllable flow rate and transparent components, it has been widely used in the separation and enrichment of CTC (Figure 1D). The Parsortix system is a microfluidic device that captures CTCs on the basis of their less deformability and larger size compared to other blood cells. Seven-point five mL of whole blood can be processed within 2 h. The main defect of this method is the difficulty of removing the leukocytes of similar size to CTCs (34) (Table 1). Another microfluidic system is ClearCell® FX1 (Clearbridge Biomedics, Singapore), which does not require preprocessing of samples and reduces the possibility of CTC loss. This device utilizes the inertial and centrifugal forces to guide the smaller RBCs and WBCs through the channel’s outer wall and the larger CTCs through the inner wall, recovering living CTCs from 1 mL of blood within 10 minutes. Equally, CTCs of the smaller sizes may flow along the channel’s outer wall, and some WBCs can be recovered in the CTC section (35). Because of the complexity of blood, the interaction between cells is not easy to control. When the sample with high cell concentration is treated, the sorting efficiency of microfluidic chip decreases. In addition, this method is solely based on the physical characteristics of cells, and human blood is a highly complex mixture of plasma, RBCs, platelets and proteins, and blood viscosity is more than three times that of water. Therefore, the chip is sometimes easy to be blocked, affecting the sorting efficiency, and the selected CTCs may have false-positive results (Table 1).
Affinity-based enrichment of CTCs
CTCs can also be isolated based on their biological properties, e.g., the presence of cell surface adhesion molecules or deficiency in hematological surface antigen markers could be utilized to distinguish CTCs from blood cells (6).
Positive selection strategy: enrichment methods with cell surface adhesion molecules
As many tumors are derived from epithelial origins, which express the EpCAM on the cell membrane to maintain cell-cell junction and adhesion, epithelial cell-specific antigens can be utilized to positively sort CTCs from billions of blood cells (36) (Figure 1E). The most widely utilized surface marker to isolate epithelial CTCs is EpCAM. Besides, cytoskeletal proteins CK8, CK18 and CK19, which are members of the cytokeratin family and also specific to epithelial cells, are the “gold standard” of cytoplasmic makers to identify epithelial CTCs (37). However, several published studies indicated that besides highly heterogenetic expression of EpCAM on tumor cells (from full expression to none) (38), many CTCs lose their epithelial marker and express mesenchymal markers during epithelial-to-mesenchymal transition (EMT). Thus, mesenchymal markers, e.g., N-cadherin (a membrane protein of the cadherin family) and vimentin (a cytoskeletal protein) are used for the detection of mesenchymal CTCs (6). Up to date, studies have shown that most CTCs are positive for both epithelial and mesenchymal markers (39). In addition, some tumor-specific markers, such as CEA, EphB4, EGFR, PSA, HER-2, MUC-1, have also been used for positive isolation of CTCs (40-43).
Immuno-magnetic beads separation using epithelial lineage markers
A widespreadly adopted method for CTC isolation using epithelial lineage markers is magnetic bead separation, where antibody-labeled ferroparticles capture CTCs in a magnetic field. Some platforms have been created to isolate and detect CTCs in previous decades. Here we would like to review a few of these commonly used devices:
CellSearch
Some platforms have been created to isolate and detect CTCs in previous decades. The most extensively used magnetic-bead based selection platform for CTC detection is the CellSearch system from Janssen Diagnostics (44-46), which is the first FDA-approved platform for CTC detection in clinical application and has been a standard method for CTC studies in various types of cancers (47-49). Potential CTCs are positively selected by using EpCAM antibody-coated magnetic beads and then characterized based on positive cytokeratins and negative CD45 (47). Using the CellSearch system, de Bono et al. showed that patients with metastatic castration-resistant PCa who had 5 or more CTCs had worse overall survival than patients with fewer than 5 cells (11.5 vs. 21.7 months) (44). Moreover, receiver operating curve analysis showed CTC enumeration is a better indicative biomarker of overall survival than PSA reduction as far as 20 weeks after starting therapy. A primary defect of this system is that CTC populations are mostly heterogeneous and several types of CTCs do not show the positivity for EpCAM (Table 1). Another pitfall of this system is that isolated CTCs lose their vitality due to fixation, which is not appropriate for subsequent culture or other functional studies (50). In view of the defects of the system, Janssen Diagnostics stopped production of the CellSearch system in 2016.
MagSweeper
As the CellSearch system has been out of production, other platforms for more sensitive and specific capture of CTCs are being developed and they display great potential. For example, the MagSweeper system made by Powell softly traps CTCs with a rod coated with anti-EpCAM antibody-labeled magnetic beads to stir a blood sample. The system has a higher rate (70%) for CTC detection in metastatic breast cancer (MBC) than the CellSearch platform. CTCs isolated by the MagSweeper strategy can keep alive (51) and are appropriate for subsequent molecular analysis, e.g., single-cell sequencing (52,53).
AdnaTest
Another technique for CTC detection based on positive selection is the AdnaTest (Qiagen). This assay method combines immunomagnetic enrichment of epithelial-derived CTCs by using anti-EpCAM and other epithelial antibodies with digital PCR for tumor-specific transcripts. Andreopoulou et al. isolated CTC using anti-EpCAM and anti-MUC1 coated magnetic beads from 55 MBC patients, and then the transcribed cDNA was subject to a commercially available molecular assay (AdnaTest BreastCancer Select) for EpCAM (also known as GA733-2), MUC-1, and HER-2. The assay is considered positive if the PCR fragment of at least one or more of the three transcripts is detected at a concentration of 0.15 ng/µL or higher. The results suggest that the AdnaTest had equivalent sensitivity to that of the CellSearch system in detecting 2 or more CTCs (54). For PCa, the assay contains primers for prostate-specific antigen (PSA), prostate-specific membrane antigen (PSMA) and the epidermal growth factor receptor (EGFR). The criteria for the sample to be positive for CTCs is also that one or more of the above transcripts are detected (55). This assay has been altered to detect the expression of the androgen receptor splice variant V7 (AR-V7) in CTCs isolated from patients with metastatic castration-resistant PCa (56). The androgen-receptor isoform encoded by splice variant 7 is deficient in the ligand-binding domain but maintains constitutively active as a transcription factor. Presence of the AR-V7 splice variant results in deregulated expression of AR-related genes (57). Antonarakis et al. found that the patients with AR-V7 positive CTCs had worse prognosis and showed resistance to the AR inhibitors abiraterone and enzalutamide (56). The researchers inferred that presence of AR-V7 positive CTCs would enable patients to benefit from non-AR targeted therapy. Scher et al. demonstrated that metastatic castration-resistant prostate cancer (mCRPC) patients harboring AR-V7-positive CTCs had better outcomes and survival with Taxanes than with the AR Inhibitors (58).
Magwire
Vermesh et al. developed a flexible magnetic wire that can achieve high local-field gradients along its entire length (59). The wire is inserted in blood vessel and captures CTCs that have been previously labelled with injected antibody-coated magnetic particles (MPs). In a live porcine model, in vivo labelling and single-pass capture of viable model CTCs could be completed within less than 10 s. The capture efficiency of the wire is comparable with to 10–80 times the amount of enriched CTCs in a 5-mL blood draw, and 500–5,000 times the enrichments achieved with the commercially available GILUPI CellCollector, which introduces an antibody-coated rod into a blood vessel for passive immunocapture of CTCs (60). Furthermore, the captured CTCs with the Magwire maintains cell viability and gene expression as unlabelled CTCs. MPs themselves have a very short half-life and is not detrimental to the organism.
Non-magnetic antigen selection using epithelial lineage markers
Another way to positively select CTCs based on cell surface markers is the application of microfluidic chips based on biochemical properties of CTCs, which can improve enrichment efficiency by increasing the surface-to-volume ratio (9). Thus far, various studies reported microfluidic devices across a great many of cancers (61-63).
IsoFlux
One remarkable example is IsoFlux by Fluxion Biosciences, which captures CTCs by using anti-EpCAM antibody-coated beads in a microfluidic chip. This enrichment technique increases the probability of catching CTCs with relatively low EpCAM expression (64).
CTC-chip
Another microfluidic system is the “CTC-chip”, which was exploited by Nagrath et al. for more efficient and selective isolation of CTCs (65). It is composed of 78,000 microposts conjugated with anti-EpCAM antibody and manipulated under preestablished laminar flow conditions without prelabeling or processing. As the blood flows through the microfluidic chip, EpCAM-positive CTCs run into the microposts and are retained (66). This strategy successfully captured CTCs in blood samples of patients with prostate, breast, lung, colon and pancreatic cancers with large CTC numbers, which reaches 99% identification rate and approximately 50% purity (65). Besides CTC-chip, several microfluidic chips based on microposts for CTC enrichment are also developed, such as geometrically enhanced differential immunocapture (GEDI)-chip (67) and Oncocee (68). A pitfall of the first-generation CTC-chip was that the sophisticated micropost design is difficult to achieve high-throughput production. An improved platform of the CTC-chip, herringbone chip (HB-chip), has been introduced. HB-chip induces the formation of microvortices generated by passive blending of blood cells to extremely increase the chance of interaction between anti-EpCAM antibodies-coated walls of the channel and CTCs (69,70). Isolated CTCs are directly used to further analysis on the device. The advantage of these chips is that many diverse tumor-specific antigens can be combined for CTC enrichment. Despite the high efficiency of these systems in isolating CTCs, acquiring intact and viable cells from the systems is still a challenge.
CMx
The CMx system is a newly developed microfluidic chip that enables effective acquisition of living CTCs. The microfluidic channels of the system are covered with a supported lipid bilayer (SLB) linked to anti-EpCAM antibody through NeutrAvidin. When the blood drawn using a syringe pump passes through microfluidic channels, the CTCs in the blood are captured by anti-EpCAM antibody while other blood cells are rinsed out by PBS. Then the CTCs are stripped with air foam, which disrupts the adhesion of the SLB to the substrate (71). The CMx platform has over 95% cell capture rate and enables living cells for subsequent characterization and in vitro culture. The system exhibits high sensitivity and specificity in distinguishing tumor patients from healthy subjects as well as patients at different stages by CTC enumeration.
NanaVelcro
NanaVelcro is another affinity-based thermo-responsive microfluidic device for CTC capture exploited by Ke et al., which is coated with 3D poly (N-isopropyl acrylamide) brushes conjugated to anti-EpCAM antibody as a thermo-responsive substrate. The substrate can enable capture and release of CTCs from the channel surface with temperature shift (72). In comparison with CellSearch, NanoVelcro has a higher capture efficiency of and enables ex vivo cultivation and functional studies (72).
Negative selection strategy: depletion methods with cell surface biomarkers
Besides the positive tumor cell markers-based affinity capture of CTC cells, CTCs can also be negatively selected based on blood cell-specific surface markers. A universe technique for WBC removal is the application of CD45 antibody-labelled magnetic beads. WBCs are depleted by mixing the blood sample with magnetic beads. (73) (Figure 1F). To date, several commercially available depletion kits are from StemCell Technologies (Vancouver, Canada), Miltenyi Biotec (Bergisch Gladbach, Germany) and ThermoFisher Scientific (Waltham, MA, USA) (74,75). One defect of the negative selection method is that not all nucleated cells in the blood express CD45. For example, the blood of healthy persons includes CD45-negative endothelial cells (73). And an even more serious pitfall of negative selection is the high loss rate of CTCs due to non-specific bulk effect, i.e., the loss of CTCs surrounded by massive clustering WBCs (Table 1).
Strategies for WBC removal are not only restricted to immunomagnetic selection. For example, the RosetteSep method, which was developed by StemCell Technologies (Vancouver, Canada), combines density gradient centrifugation with negative enrichment (76-78). Both WBCs and RBCs are cross-linked to form clusters by utilizing bispecific antibodies targeting CD45, CD66b and glycophorin and then depleted through density gradient separation. The CD45/CD66b-expressing cells are distributed in the lower compartment and the CD45/CD66b-deficient nucleated cells and CTCs accumulates in the interface between the plasma and the density medium.
Lin et al. successfully developed a novel approach integrating subtraction enrichment and immunostaining-FISH (SE-iFISH), which effectively depletes WBCs and RBCs and detects nonhematopoietic CTCs, circulating tumor microemboli (CTM) and disseminated tumor cells (DTCs) based on chromosomal aneuploid derived from a wide variety of types of solid tumor regardless of CK or EpCAM expression and CTC size variation (38,79,80). Most of normal residual fibroblasts and endothelial cells following enrichment are diploid, which could be distinguished from those with abnormal aneuploid chromosome. Remaining endothelial cells were further pinpointed by CD31 staining. The CTCs enriched with this strategy are viable and suitable for extended culture and extra functional study. Compared with CellSearch (54.8%), SE-iFISH had a higher positive rate of CTCs (90.5%) from the same population of gastric cancer patients (81). Similarly, high sensitivity for CTC detection by SE-iFISH was also observed on lung (92%) and esophageal (87%) cancer patients (79). In addition, the CTCs with diverse subtypes may indicate distinct clinical significance. In situ phenotyping and karyotyping analysis of highly heterogeneous subpopulations of CTC/DTC from gastric cancer patients (in situ PK CTC or DTC) performed using SE-iFISH showed that among CK18 negative CTCs, trisomy chromosome 8 CTCs may exhibit intrinsic resistance to the chemotherapeutic agent cisplatin, while tetra- and pentasomy subtype developed acquired resistance (81). Similarly, both cisplatin-sensitive and insensitive CTCs in gastric neuroendocrine cancer PDX mice could be characterized and classified by CK18-iFISH (82).
Multi-marker immune-magnetic negative depletion enrichment of CTCs (MINDEC), a newly improved negative depletion strategy, is based on various markers including CD45 (pan-leukocyte), CD16 (natural killer cells and neutrophil granulocytes), CD19 (B-cells), CD163 (monocytes and macrophages), and CD235a (RBCs) to remove blood cells and capture CTCs from blood samples (83). The strategy exhibits an average recovery of 82%±10% and high removal (437±350 residual WBCs). The MINDEC strategy detects CTCs in 71% of metastatic pancreatic cancer patients. One defect of this method is that it is possible that aggregates of CTCs enclosed with WBCs, e.g., CTC microembolisms (CTMs) may be depleted from the CTCs enriched with this strategy.
In addition to specific antibodies, affinity-binding enrichment techniques use polypeptide (84) or aptamer (85,86) (a single-stranded DNA or RNA molecule that binds specifically to CTC or leukocyte surface antigen) instead of antibodies to achieve positive or negative enrichment.
In brief, positive and negative enrichment techniques based on the expression of epithelial cell markers are widely used in CTC detection. A defect of these methods is that CTCs which exhibits low or no EpCAM expression during the EMT could not be detected. Thus, these approaches may frequently produce false-negative results due to the presence of such CTCs. The methods based on physical properties eliminate the above problem. However, as there are many similar physical properties between blood cells and CTCs, this class of methods often have a relatively high false-positive rate. The specificity and sensitivity of CTC enrichment methods should be balanced for further development in the future.
A combination of multiple properties for CTC isolation
The acquisition of CTCs with higher recovery and purity necessitates the combined application of techniques for CTC detection. Researchers from Massachusetts General Hospital developed the third-generation microfluidic chip, CTC-iChip, which relies on a combination of multiple properties including size exclusion and affinity-based immuno-magnetic beads selection to get CTCs with higher purity from blood samples (87). First, RBCs, platelets, plasma proteins and free magnetic beads are separated from WBCs and CTCs by the microspots of the CTC-iChip based on hydrodynamic cell size. Then the principles of inertial focusing were applied to order nucleated cells within the microfluidic channel. Finally, residual WBCs were separated from CTCs by using pre-mixed CD45 and CD15 immuno-magnetic beads. The CTC-iChip has achieved a mean cell recovery of 95% in multiple cell lines and predicted 90% of tumor recurrence in prostate tumor patients through CTC enumeration. However, it is not suitable for CTCs with small diameters (<8 µm) (88). Equally, GEDI-chip, which was worked out by Kirby et al., is characterized by the combination of size-based exclusion and affinity-based selection for the enrichment of the PSMA-positive CTCs from blood samples of castration-resistant prostate tumor patients (67). The number of CTCs isolated with GEDI are 200 to 400 folds of that achieved using CellSearch in the same patient, demonstrating a considerable enhancement in the efficiency of CTC isolation. In addition, NanoFlares are nanoconstructs that could detect intracellular mRNAs of viable cells at the single-cell level. The NanoFlares, with the aid of flow cytometry, can fluorescently detect genetic markers of CTCs in the blood. They enable the detection of as few as 100 viable human breast cancer cells per mL of blood and subsequent cultivation of isolated cells, as well as the identification of the CTCs in a murine model of MBC. The NanoFlares is the first method for simultaneously labeling, isolating and genetically characterizing viable CTCs and may provide a novel tool for clinicians to evaluate recurrent risk and therapy response (89).
Perspective and conclusions
The scarcity of CTCs in the blood has pushed forward the development of more and more systems and techniques for the enrichment and characterization of CTCs. In this review, we summarized the technical superiorities and defects of the available CTC isolation and detection methods. Microfluidic chip technology has certain advantages in cell sorting due to its own characteristics, including small chip size, high speed, high flux, convenient operation, low sample and reagent consumption, easy integration of multi-purpose functional components on the chip. With the development of nanotechnology, microfluidic chips modified with functionalized nanomaterials are widely used in the enrichment and detection of CTCs. Antibody-linked functional nanoparticles can provide a larger contact surface area for CTC-antibody binding. Aptamers, including single-stranded ribonucleic acids and deoxyribonucleic acids, are molecules that have high affinity for, and can selectively bind to, a specific target, e.g., ions, small molecules, cells (90). The aptamer can provide specific CTC targets, so the application of microfluidic chip may be towards the development of new CTC capture probes, such as nucleic acid aptamer probes. Microfluidic chip technology has been widely used in CTC sorting, and is expected to become one of the primary tools for CTC enrichment and detection in the future. However, it is quite challenging to come up with an efficient strategy with high sensitivity and specificity that can enrich the entire CTCs. In addition, different techniques of CTC capture may have different consequences. Thus, the combined application of these strategies may be a trend for most primary CTC detection techniques. For example, CTC-iChip, the third-generation chip, enriches CTC with deterministic lateral displacement, inertial focusing and immunomagnetic beads. Furthermore, no ideal marker is expressed on every CTC and shows constant expression with the progression of the disease, in view of the heterogeneity of CTCs. The use of large panels of biomarkers, including epithelial biomarkers, mesenchymal markers or tumor-specific biomarkers, is required for total CTC detection in blood samples. Although CTC detection still faces various challenges, progresses are constantly being made in CTC study. Detection of CTCs still play an important role for further screen potential markers for early tumor diagnosis, explore new therapeutic targets, and develop promising strategies for tumor prevention and therapy. The technology for detection of CTCs in patients still has bright perspectives. Cancer patients will greatly benefit from any improvement and breakthrough of CTC detection technologies.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.01.17). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ashworth TR. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust Med J 1869;14:146-7.
- Watanabe S. The metastasizability of tumor cells. Cancer 1954;7:215-23. [Crossref] [PubMed]
- Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer 2009;9:302-12. [Crossref] [PubMed]
- Lloyd JM, McIver CM, Stephenson SA, et al. Identification of early-stage colorectal cancer patients at risk of relapse post-resection by immunobead reverse transcription-PCR analysis of peritoneal lavage fluid for malignant cells. Clin Cancer Res 2006;12:417-23. [Crossref] [PubMed]
- Hüsemann Y, Geigl JB, Schubert F, et al. Systemic spread is an early step in breast cancer. Cancer Cell 2008;13:58-68. [Crossref] [PubMed]
- Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer 2014;14:623-31. [Crossref] [PubMed]
- Meng S, Tripathy D, Frenkel EP, et al. Circulating tumor cells in patients with breast cancer dormancy. Clin Cancer Res 2004;10:8152-62. [Crossref] [PubMed]
- Zhe X, Cher ML, Bonfil RD. Circulating tumor cells: finding the needle in the haystack. Am J Cancer Res 2011;1:740-51. [PubMed]
- Arya SK, Lim B, Rahman AR. Enrichment, detection and clinical significance of circulating tumor cells. Lab Chip 2013;13:1995-2027. [Crossref] [PubMed]
- Seal SH. Silicone flotation: a simple quantitative method for the isolation of free-floating cancer cells from the blood. Cancer 1959;12:590-5. [Crossref] [PubMed]
- Hou HW, Warkiani ME, Khoo BL, et al. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci Rep 2013;3:1259. [Crossref] [PubMed]
- Pösel C, Möller K, Fröhlich W, et al. Density gradient centrifugation compromises bone marrow mononuclear cell yield. PLoS One 2012;7:e50293. [Crossref] [PubMed]
- Gertler R, Rosenberg R, Fuehrer K, et al. Detection of circulating tumor cells in blood using an optimized density gradient centrifugation. Recent Results Cancer Res 2003;162:149-55. [Crossref] [PubMed]
- Rosenberg R, Gertler R, Friederichs J, et al. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry 2002;49:150-8. [Crossref] [PubMed]
- Danova M, Torchio M, Mazzini G. Isolation of rare circulating tumor cells in cancer patients: technical aspects and clinical implications. Expert Rev Mol Diagn 2011;11:473-85. [Crossref] [PubMed]
- Szczerba BM, Castro-Giner F, Vetter M, et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 2019;566:553-7. [Crossref] [PubMed]
- Chen JF, Ho H, Lichterman J, et al. Subclassification of prostate cancer circulating tumor cells by nuclear size reveals very small nuclear circulating tumor cells in patients with visceral metastases. Cancer 2015;121:3240-51. [Crossref] [PubMed]
- Williams ES, Rodriguez-Bravo V, Chippada-Venkata U, et al. Generation of prostate cancer patient derived xenograft models from circulating tumor cells. J Vis Exp 2015;53182. [PubMed]
- Ma YC, Wang L, Yu FL. Recent advances and prospects in the isolation by size of epithelial tumor cells (ISET) methodology. Technol Cancer Res Treat 2013;12:295-309. [Crossref] [PubMed]
- Zheng S, Lin H, Liu JQ, et al. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J Chromatogr A 2007;1162:154-61. [Crossref] [PubMed]
- Ilie M, Szafer-Glusman E, Hofman V, et al. Expression of MET in circulating tumor cells correlates with expression in tumor tissue from advanced-stage lung cancer patients. Oncotarget 2017;8:26112-21. [Crossref] [PubMed]
- Dolfi SC, Chan LL, Qiu J, et al. The metabolic demands of cancer cells are coupled to their size and protein synthesis rates. Cancer Metab 2013;1:20. [Crossref] [PubMed]
- Alunni-Fabbroni M, Sandri MT. Circulating tumour cells in clinical practice: Methods of detection and possible characterization. Methods 2010;50:289-97. [Crossref] [PubMed]
- Kim MS, Sim TS, Kim YJ, et al. SSA-MOA: a novel CTC isolation platform using selective size amplification (SSA) and a multi-obstacle architecture (MOA) filter. Lab Chip 2012;12:2874-80. [Crossref] [PubMed]
- Guck J, Schinkinger S, Lincoln B, et al. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys J 2005;88:3689-98. [Crossref] [PubMed]
- Swaminathan V, Mythreye K, O'Brien ET, et al. Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer Res 2011;71:5075-80. [Crossref] [PubMed]
- Zhang W, Kai K, Choi DS, et al. Microfluidics separation reveals the stem-cell-like deformability of tumor-initiating cells. Proc Natl Acad Sci U S A 2012;109:18707-12. [Crossref] [PubMed]
- Shaw Bagnall J, Byun S, Begum S, et al. Deformability of tumor cells versus blood cells. Sci Rep 2015;5:18542. [Crossref] [PubMed]
- Riahi R, Gogoi P, Sepehri S, et al. A novel microchannel-based device to capture and analyze circulating tumor cells (CTCs) of breast cancer. Int J Oncol 2014;44:1870-8. [Crossref] [PubMed]
- Gogoi P, Sepehri S, Zhou Y, et al. Development of an automated and sensitive microfluidic device for capturing and characterizing circulating tumor cells (CTCs) from clinical blood samples. PLoS One 2016;11:e0147400. [Crossref] [PubMed]
- Gupta V, Jafferji I, Garza M, et al. ApoStream(™), a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics 2012;6:24133. [Crossref] [PubMed]
- Poklepovic AS, Wan W, Wu W, et al. ApoStream, an antibody-independent platform, compared to CellSearch for enumeration of circulating tumor cells (CTCs) in patients with metastatic prostate cancer. J Clin Oncol 2012;30:abstr e21058.
- Li P, Mao Z, Peng Z, et al. Acoustic separation of circulating tumor cells. Proc Natl Acad Sci U S A 2015;112:4970-5. [Crossref] [PubMed]
- Xu L, Mao X, Imrali A, et al. Optimization and evaluation of a novel size based circulating tumor cell isolation system. PLoS One 2015;10:e0138032. [Crossref] [PubMed]
- Lee Y, Guan G, Bhagat AA. ClearCell(R) FX, a label-free microfluidics technology for enrichment of viable circulating tumor cells. Cytometry A 2018;93:1251-4. [Crossref] [PubMed]
- Pantel K, Alix-Panabieres C. Real-time liquid biopsy in cancer patients: fact or fiction? Cancer Res 2013;73:6384-8. [Crossref] [PubMed]
- Lu SH, Tsai WS, Chang YH, et al. Identifying cancer origin using circulating tumor cells. Cancer Biol Ther 2016;17:430-8. [Crossref] [PubMed]
- Lin PP. Integrated EpCAM-independent subtraction enrichment and iFISH strategies to detect and classify disseminated and circulating tumors cells. Clin Transl Med 2015;4:38. [Crossref] [PubMed]
- Armstrong AJ, Marengo MS, Oltean S, et al. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res 2011;9:997-1007. [Crossref] [PubMed]
- Ghazani AA, Castro CM, Gorbatov R, et al. Sensitive and direct detection of circulating tumor cells by multimarker µ-nuclear magnetic resonance. Neoplasia 2012;14:388-95. [Crossref] [PubMed]
- Pecot CV, Bischoff FZ, Mayer JA, et al. A novel platform for detection of CK+ and CK- CTCs. Cancer Discov 2011;1:580-6. [Crossref] [PubMed]
- Yu M, Bardia A, Wittner BS, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013;339:580-4. [Crossref] [PubMed]
- Santana SM, Liu H, Bander NH, et al. Immunocapture of prostate cancer cells by use of anti-PSMA antibodies in microdevices. Biomed Microdevices 2012;14:401-7. [Crossref] [PubMed]
- de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9. [Crossref] [PubMed]
- Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:3213-21. [Crossref] [PubMed]
- Hayes DF, Cristofanilli M, Budd GT, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res 2006;12:4218-24. [Crossref] [PubMed]
- Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 2004;10:6897-904. [Crossref] [PubMed]
- Hall C, Karhade M, Laubacher B, et al. Circulating tumor cells after neoadjuvant chemotherapy in stage I-III triple-negative breast cancer. Ann Surg Oncol 2015;22:S552-8. [Crossref] [PubMed]
- Zhou J, Dong F, Cui F, et al. The role of circulating tumor cells in evaluation of prognosis and treatment response in advanced non-small-cell lung cancer. Cancer Chemother Pharmacol 2017;79:825-33. [Crossref] [PubMed]
- Andree KC, van Dalum G, Terstappen LW. Challenges in circulating tumor cell detection by the CellSearch system. Mol Oncol 2016;10:395-407. [Crossref] [PubMed]
- Talasaz AH, Powell AA, Huber DE, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci U S A 2009;106:3970-5. [Crossref] [PubMed]
- Powell AA, Talasaz AH, Zhang H, et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One 2012;7:e33788. [Crossref] [PubMed]
- Deng G, Krishnakumar S, Powell AA, et al. Single cell mutational analysis of PIK3CA in circulating tumor cells and metastases in breast cancer reveals heterogeneity, discordance, and mutation persistence in cultured disseminated tumor cells from bone marrow. BMC Cancer 2014;14:456. [Crossref] [PubMed]
- Andreopoulou E, Yang LY, Rangel KM, et al. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect versus Veridex CellSearch system. Int J Cancer 2012;130:1590-7. [Crossref] [PubMed]
- Todenhöfer T, Hennenlotter J, Feyerabend S, et al. Preliminary experience on the use of the Adnatest® system for detection of circulating tumor cells in prostate cancer patients. Anticancer Res 2012;32:3507-13. [PubMed]
- Antonarakis ES, Lu C, Wang H, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014;371:1028-38. [Crossref] [PubMed]
- Hu R, Dunn TA, Wei S, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res 2009;69:16-22. [Crossref] [PubMed]
- Scher HI, Lu D, Schreiber NA, et al. Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol 2016;2:1441-9. [Crossref] [PubMed]
- Vermesh O, Aalipour A, Ge TJ, et al. An intravascular magnetic wire for the high-throughput retrieval of circulating tumour cells in vivo. Nat Biomed Eng 2018;2:696-705. [Crossref] [PubMed]
- Saucedo-Zeni N, Mewes S, Niestroj R, et al. A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. Int J Oncol 2012;41:1241-50. [PubMed]
- Murlidhar V, Zeinali M, Grabauskiene S, et al. A radial flow microfluidic device for ultra-high-throughput affinity-based isolation of circulating tumor cells. Small 2014;10:4895-904. [Crossref] [PubMed]
- Thege FI, Lannin TB, Saha TN, et al. Microfluidic immunocapture of circulating pancreatic cells using parallel EpCAM and MUC1 capture: characterization, optimization and downstream analysis. Lab Chip 2014;14:1775-84. [Crossref] [PubMed]
- Park JM, Kim MS, Moon HS, et al. Fully automated circulating tumor cell isolation platform with large-volume capacity based on lab-on-a-disc. Anal Chem 2014;86:3735-42. [Crossref] [PubMed]
- Harb W, Fan A, Tran T, et al. Mutational analysis of circulating tumor cells using a novel microfluidic collection device and qPCR assay. Transl Oncol 2013;6:528-38. [Crossref] [PubMed]
- Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007;450:1235-9. [Crossref] [PubMed]
- Sequist LV, Nagrath S, Toner M, et al. The CTC-chip: an exciting new tool to detect circulating tumor cells in lung cancer patients. J Thorac Oncol 2009;4:281-3. [Crossref] [PubMed]
- Kirby BJ, Jodari M, Loftus MS, et al. Functional characterization of circulating tumor cells with a prostate-cancer-specific microfluidic device. PLoS One 2012;7:e35976. [Crossref] [PubMed]
- Kalinsky K, Mayer JA, Xu X, et al. Correlation of hormone receptor status between circulating tumor cells, primary tumor, and metastasis in breast cancer patients. Clin Transl Oncol 2015;17:539-46. [Crossref] [PubMed]
- Stott SL, Lee RJ, Nagrath S, et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci Transl Med 2010;2:25ra3. [Crossref] [PubMed]
- Stott SL, Hsu CH, Tsukrov DI, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A 2010;107:18392-7. [Crossref] [PubMed]
- Chen JY, Tsai WS, Shao HJ, et al. Sensitive and specific biomimetic lipid coated microfluidics to isolate viable circulating tumor cells and microemboli for cancer detection. PLoS One 2016;11:e0149633. [Crossref] [PubMed]
- Ke Z, Lin M, Chen JF, et al. Programming thermoresponsiveness of NanoVelcro substrates enables effective purification of circulating tumor cells in lung cancer patients. ACS Nano 2015;9:62-70. [Crossref] [PubMed]
- Yang L, Lang JC, Balasubramanian P, et al. Optimization of an enrichment process for circulating tumor cells from the blood of head and neck cancer patients through depletion of normal cells. Biotechnol Bioeng 2009;102:521-34. [Crossref] [PubMed]
- Lee MY, Lufkin T. Development of the "Three-step MACS": a novel strategy for isolating rare cell populations in the absence of known cell surface markers from complex animal tissue. J Biomol Tech 2012;23:69-77. [Crossref] [PubMed]
- Wu Y, Deighan CJ, Miller BL, et al. Isolation and analysis of rare cells in the blood of cancer patients using a negative depletion methodology. Methods 2013;64:169-82. [Crossref] [PubMed]
- Reinholz MM, Kitzmann KA, Tenner K, et al. Cytokeratin-19 and mammaglobin gene expression in circulating tumor cells from metastatic breast cancer patients enrolled in North Central Cancer Treatment Group trials, N0234/336/436/437. Clin Cancer Res 2011;17:7183-93. [Crossref] [PubMed]
- Sun YF, Xu Y, Yang XR, et al. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology 2013;57:1458-68. [Crossref] [PubMed]
- Guo W, Sun YF, Shen MN, et al. Circulating tumor cells with stem-like phenotypes for diagnosis, prognosis, and therapeutic response evaluation in hepatocellular carcinoma. Clin Cancer Res 2018;24:2203-13. [Crossref] [PubMed]
- Ge F, Zhang H, Wang DD, et al. Enhanced detection and comprehensive in situ phenotypic characterization of circulating and disseminated heteroploid epithelial and glioma tumor cells. Oncotarget 2015;6:27049-64. [Crossref] [PubMed]
- Lin PP, Gires O, Wang DD, et al. Comprehensive in situ co-detection of aneuploid circulating endothelial and tumor cells. Sci Rep 2017;7:9789. [Crossref] [PubMed]
- Li Y, Zhang X, Ge S, et al. Clinical significance of phenotyping and karyotyping of circulating tumor cells in patients with advanced gastric cancer. Oncotarget 2014;5:6594-602. [Crossref] [PubMed]
- Jiang J, Wang DD, Yang M, et al. Comprehensive characterization of chemotherapeutic efficacy on metastases in the established gastric neuroendocrine cancer patient derived xenograft model. Oncotarget 2015;6:15639-51. [PubMed]
- Lapin M, Tjensvoll K, Oltedal S, et al. MINDEC-an enhanced negative depletion strategy for circulating tumour cell enrichment. Sci Rep 2016;6:28929. [Crossref] [PubMed]
- Yue C, Jiang Y, Li P, et al. Dynamic change of PD-L1 expression on circulating tumor cells in advanced solid tumor patients undergoing PD-1 blockade therapy. Oncoimmunology 2018;7:e1438111. [Crossref] [PubMed]
- Song Y, Zhu Z, An Y, et al. Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal Chem 2013;85:4141-9. [Crossref] [PubMed]
- Wang DL, Song YL, Zhu Z, et al. Selection of DNA aptamers against epidermal growth factor receptor with high affinity and specificity. Biochem Biophys Res Commun 2014;453:681-5. [Crossref] [PubMed]
- Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med 2013;5:179ra47. [Crossref] [PubMed]
- Karabacak NM, Spuhler PS, Fachin F, et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc 2014;9:694-710. [Crossref] [PubMed]
- Halo TL, McMahon KM, Angeloni NL, et al. NanoFlares for the detection, isolation, and culture of live tumor cells from human blood. Proc Natl Acad Sci U S A 2014;111:17104-9. [Crossref] [PubMed]
- Gao T, Luo Y, Li W, et al. Progress in the isolation of aptamers to light-up the dyes and the applications. Analyst 2020;145:701-18. [PubMed]


