
5-aza-2’-deoxycytidine (DAC) treatment induces the MAGE-A10 expression and improves the cytotoxicity of MAGE-A10-specific CTLs in lung cancer cells
Introduction
Lung cancer is one of the most common malignant tumors. In most cases, lung cancer is diagnosed at an advanced disease stage with metastases to other organ sites resulting in a poor survival rate. The treatment of lung cancer involves a combination of conventional methods, including chemotherapy, radiotherapy, and surgery as well as immune- and targeted therapy. However, the clinical prognosis remains poor.
Melanoma-associated antigen A (MAGE-A) belongs to the MAGE group of well-characterized members of the cancer/testis antigen (CTA) family, that are expressed in normal cells of the testis, placenta and in fetal tissue (1,2). Furthermore, it was recently reported that MAGE-A family proteins contribute to various malignancies (3). MAGE-A10 is a subtype of the MAGE-A family. Previous studies have reported that MAGE-A10 is frequently highly expressed in the tumor cells in skin and urothelial malignancies (4,5). However, its role in lung cancer remains unclear.
In recent years, immunotherapy has shown its dominant advantage and has therefore become a promising therapeutic modality for various types of cancers (6,7). However the success of immunotherapy is influenced by various biological processes including tumor immune escape, one of the major reasons for the failure of tumor immunotherapy (8). Tumor immune escape refers to the phenomenon by which tumor cells escape the recognition and attack by the immune system through various mechanisms, such as inhibiting the differentiation of cytotoxic T lymphocytes (CTLs), that reduces the recognition and attack of the tumor cells by the immune system, thereby, surviving immune elimination (9,10). Several lines of evidence have revealed that the MAGE-A family can promote the responsiveness of specific CTLs (11). However, whether high expression of MAGE-A10 can specifically enhance the effect of the immune response of CTLs in lung cancer is unknown (12).
Epigenetic-modifying drugs can enhance the expression of tumor antigens and improve the cytotoxicity of antigen-specific T cells (13). 5-aza-2’-deoxycytidine (DAC), a DNA methyltransferase inhibitor (DNMTI) considered to be an epigenetic-modifying drug, enhanced the expression of MAGE-A10 (14,15). Kaplan-Meier plotter (www.kmplot.com) is an online database that integrates gene expression and clinical data, especially survival information. This online database can be used to obtain the survival curve for specific types of cancers, such as lung cancer, ovarian cancer, gastric cancer, and breast cancer.
Therefore, in our study we use the online database to explore the prognostic role of MAGE-A10 expression levels in lung cancer patients. Subsequently, DAC was used to induce the expression of MAGE-A10 in lung cancer cells and the cytotoxicity of MAGE-A10-specific CTLs on lung cancer cells pretreated by DAC was evaluated.
Methods
Date mining of Kaplan-Meier Plotter dataset and statistical analysis
The overall survival (OS) time is defined from diagnosis to death or the last follow-up. The post-progression survival (PPS) is defined as the time after the patient’s tumor progression. The first progression (FP) is defined from diagnosis to FP. Survival curves were obtained from the online database of the Kaplan-Meier Plotter (http://www.kmplot.com). Data were analyzed using SPSS 19.0 statistical software (SPSS Inc., IL, and USA). P<0.05 was considered statistically significant.
Immunohistochemistry (IHC) analysis
Immunohistochemistry analysis was performed on formalin-fixed, paraffin-embedded samples. MAGE-A10 immune staining was assessed on whole tissue sections. Antigen retrieval was performed by microwaving the array in sodium citrate buffer for 10 min. Immunohistochemical staining was performed according to a previously described protocol. The sections were incubated with a mouse antibody against human MAGE-A10 (clone 6C1, Invitrogen, Carlsbad, CA) at a dilution of 1:500 overnight at 4 °C, followed by incubation with an HRP-conjugated secondary antibody and substrate. The mean number of immunopositive cells in the samples was determined in five fields-of-view. Immunohistochemical staining was then evaluated according to the Friedrich immunoreactivity score (IRS) which grades immunohistochemical staining in two categories. Firstly, based on the percentage of stained cells: <1% (score =0); 1–25% (score =1); 25–50% (score =2); 51–80% (score =3); >80% (score =4). Secondly, based on the staining intensity or depth of the color: no staining (score =0); buff (score =1); darker buff (score =2); tan (score =3). Finally, the scores were multiplied to obtain IRS scores, and described as either low (score 0–3, −/+), moderate (score 4–7, ++) or high (score >7, +++). The immuno reactivity scores for MAGE-A10 was presented as either “negative” or “positive,” with positive including moderate and high reactions scores. The percentage of stained positive cells and staining depth were scored by three independent observers.
Cell lines
Human lung cancer cell lines (H1975 and A549) were purchased from the cell bank of Chinese Academy of Sciences (Shanghai, China). Primary lung cancer cells (L228, L329, and L419) were collected from patients in our hospital. All these cells were used as target cells and cultured in RPMI-1640 (GIBCO, Grand Island, NY, USA), containing 1% Penicillin-Streptomycin (100 U/mL penicillin and 100 µg/mL streptomycin) and 10% fetal bovine serum (FBS, HyClone, Logan, UT, USA) at 37 °C with 5% CO2.
DAC treatment
5-aza-2’-deoxycytidine (DAC), a DNA methyltransferase inhibitors (DNMTis) considered as an epigenetic-modifying drug, could enhance the expression of MAGE-A10 (12). DAC was dissolved in distilled water and prepared at 3 doses (25, 50, and 100 µg). Lung cancer cells either untreated or pretreated were treated with DAC for 72 h. Subsequently, the cells were harvested for the preparation of protein lysates and RNA isolation. Then, RT-PCR was used to explore gene expression in the lung cell lines and western blot analysis was used to study the protein expression.
Peptide induced CTLs
MAGE-A10 antigenic peptide (sequence: SLLKFLAKV) was prepared and synthesized as described previously (Beijing Huaxia Ocean Technology Co., Ltd. Haiding district, Beijing). Dendritic cells (DCs)and CD8+T cells were separated and purified from peripheral blood mononuclear cells (PBMCs) obtained from healthy volunteers(with informed consent)by Ficoll-Hypaque density gradient centrifugation. DCs were cultured for seven days with 100 ng/mL GM-CSF (granulocyte-macrophage colony stimulating factor) and 50 ng/mL IL-4 (Peprotech). Tumor necrosis factor (TNF-α, 20 ng/mL; Peprotech) and maturation agent lipopolysaccharide (LPS, 10 ng/mL; Peprotech) were added to the culture for 24 h on day five. Then the DCs were pulsed with MAGE-A10 antigenic peptides (10 µM) for 5 h. Subsequently, the autologous CD8+T cells were stimulated with the peptide-loaded DCs. MAGE-A10-specific CTLs were harvested after 2 weeks of stimulation.
Real-time PCR
Extraction of total RNA, complementary DNA (cDNA) synthesis and RT-PCR analyses were performed as described previously. Total RNA was extracted from the H1975, A549, L228, L329, and L419 cells with TRIzol reagent (Invitrogen, Shanghai, China) according to the manufacturer’s protocol. cDNA was generated from the isolated total RNA using random primers and reverse transcription with Superscript II (Invitrogen). The primers were obtained from Takara (Dalian, China). The thermocycler conditions were as follows: initial denaturation at 50 °C for 2 minutes and then 95 °C for 10 minutes; then followed by a two-step PCR program of 95 °C for 15 seconds and 60 °C for 1 minute repeated for 35 cycles and a final elongation step at 72 °C for ten minutes. The RT-PCR program was performed on a Mx4000 system (Applied Biosystems, Foster City, CA), which was used to collect data and for quantitative analysis. The expression level of MAGE-A10 mRNA was shown as fold change relative to the internal control. The primers utilized for the quantitative RT-PCR assays are shown in Table 1 (16).
Table 1
| Protein | Forward | Reverse |
|---|---|---|
| MAGE-A10 | 5'-CACAGAGCAGCACTGAAGGAG-3' | 5'-CTGGGTAAAGACTCACTGTCTGG-3' |
| β-Actin | 5'-CTACGTCGCCCTGGACTTCGAGC-3' | 5'-GATGGAGCCGCCGATCCACACGG-3' |
Western blot analysis
Total proteins were extracted from each cell line using RIPA buffer (Beyotime Institute of Biotechnology, China) supplemented with a protease inhibitor, and the protein concentration was quantified using BCA Protein Assay Kit (Thermo Scientific). Equal amounts of protein were separated by 10% SDS-PAGE and transferred to PVDF membranes at 300 mA for 2 h. The membranes were blocked in Tris-buffered saline with Tween-20 (100 mM NaCl, 50 mM Tris-HCI, pH 8.0 and 0.1% Tween 20) containing 5% non-fat dry milk for 2 h at 37 °C, followed by overnight incubation at 4 °C with the primary antibodies. The primary antibodies used were anti-MAGE-A10 (Abcam, Britain) and antitoxin (Santa Cruz). After washes, horseradish peroxidase (HRP)-coupled secondary anti-mouse or anti-rabbit antibodies were incubated with the membranes and agitated on a shaker for 2 h at room temperature. The target proteins were visualized using an enhanced chemiluminescence (ECL) detection system (Thermo Scientific). The molecular weight of MAGE-A10 was 72 kDa. The band densities were quantitated using the Quantity One software (BIO-RAD).
Enzyme-linked immunospot assay
Enzyme-linked immunospot (ELISPOT) was used to detect the capacity of CTLs induced by MAGE-A10-specific peptide. ELISA was performed according to the manufacturer’s instructions (Shanghai ruizhi Bio-pharmaceutical Technology Co., Ltd.) A 96-well plate was coated with anti-human IFN-γ. Cell suspension in 100 µL volume containing about 1×105 T cells were added to each well were set for each group. Irrelevant peptides and phytohemagglutinin were used as negative and positive controls, respectively. The 96-well plate were covered and incubated in a 37 °C CO2 incubator for 15–20 h without any agitation or movement. A total of 100 µL of BCIP/NBT was added per well and the plate was further incubated. The purple spots visualized in the PVDF wells represent cells producing the cytokine. The spots were counted manually under a microscope or by the ELISA assay system.
Cell counting kit (CCK-8) assay
CCK-8 assay was used to determine the cytotoxic activity of peptide-specific CTLs and CD8+ T lymphocytes untreated with peptide. The lung cancer cells (104/well) were suspended in 50 mL RPMI medium containing 10% fetal calf serum and plated in the wells of a 96-well round-bottom plate for 24 h. The attached cells were divided into four groups. Two of the groups were treated with DAC for 3 days (Group 1and 2). And others were left untreated (Group 3 and 4). Group 1 cells received MAGE-A10-specific CTLs + DAC. Group 2 cells received only DAC. Group 3 cells receivedMAGE-A10-specific CTLs. Group 4 cells remained untreated. The four groups were treated for 1 day according to a specific E:T ratio (10:1,20:1, 40:1). We add 10 mL of the CCK-8 solution to each well after 24 h. The absorbance was read at 450 nm after 3 h of incubation and the cytotoxic capacity of the CTL was measured. The percentage of lysis of the target cells was calculated as follows: . Subsequently, the supernatant was collected to detect the amount of IFN-γ.
Results
MAGE-A10is an unfavorable prognostic indicator in human lung cancer
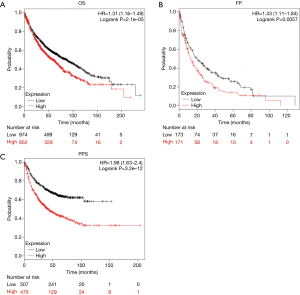
Data pertaining to lung cancer patients with complete clinical records were analyzed from the online Kaplan-Meier plotter database. Higher expression of MAGE-A10 (split patients by median expression) was found to be negatively associated with OS [1,926 patients fit the OS analysis, HR =1.31, Plogrank =2.1e-05, 95% CI (1.16–1.49)] and PPS [344 patients fit the PPS analysis, HR =1.43, Plogrank =0.0057, 95% CI (1.11–1.84)], and FP [982 patients fit the FP analysis, HR =1.98, Plogrank =3.2e-12, 95% CI (1.63–2.4)]. MAGE-A10 protein expression was identified as an unfavorable prognostic factor for patients with lung cancer (Figure 1).
MAGE-A10 was highly expressed in patient with lung cancer
The expression of MAGE-A10 was analyzed in 103 lung cancer tissues using IHC. MAGE-A10 expression was found in 57/103 (55.34%) lung cancer tissues and 4/30 (13.3%) non-cancerous healthy tissues. A significant difference was found in the expression level of MAGE-A10 protein between lung cancer tissues and adjacent tissues (P value <0.05). Based on the survival time of the group, the results indicated an association between MAGE-A10 expression and prognosis of the 103 patients with lung cancer. Patients had a poorer prognosis when the expression of MAGE-A10 was high, further patients had a better prognosis when the expression of MAGE-A10 was low (Figure 2, Table 2). This data demonstrates that the expression of MAGEA-10 is significantly associated with the prognosis of lung cancer.
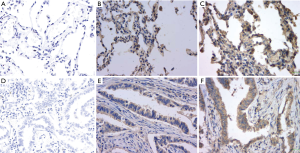
Table 2
| Tissue | MAGE-A10 | P | ||
|---|---|---|---|---|
| −/+ (%) | ++ (%) | +++ (%) | ||
| Tumor tissue | 5 (22.7) | 9 (40.9) | 8 (36.4) | <0.05 |
| Adjacent tissue | 6 (24.0) | 9 (36.0) | 10 (40.0) | |
*, P<0.05. Chi-squared test was used.
The expression of MAGE-A10 in different cancer cells
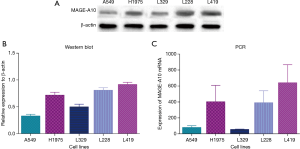
Western blot analysis was used to detect the expression of MAGE-A10 in different cancer cells. Furthermore, we used RT-PCR to detect the mRNA expression level of MAGE-A10 in the cell lines (Figure 3).
The expression of MAGE-A10 in lung cancer cells following treatment with DAC
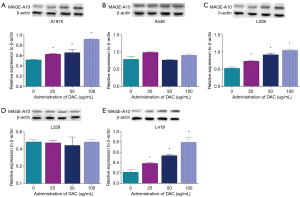
DAC, DNA methyltransferase inhibitors (DNMTis) was chosen to induce the expression of MAGE-A10. RT-PCR and western blotting were used to detect the effect of DAC on select lung cancer cells. The expression of MAGE-A10was significantly increased in DAC treated L419, L228 and H1975cells in a dose-dependent manner compared to the untreated cells. Whereas, there was no statistical significance in the L329 and A549 cells. Therefore we selected L419, L228 and H1975 as candidate target cells to investigate the ability of MAGE-A10 in killing effect of CTLs (Figure 4).
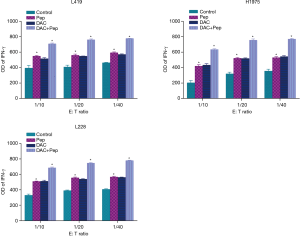
MAGE-A10 antigenic peptide significantly inducedMAGE-A10-specific CTLs
CD8+ T cells were stimulated with MAGE-A10-pulsed DCs. CCK-8 assay and IFN-γ release ELISA assay was used to test the capacity of inducing MAGE-A10-specific CTLs by MAGE-A10-pulsed DCs and to assess the cytotoxic activity of MAGE-A10-specific CTLs. CCK-8 assays showed that the MAGE-A10-specific CTLs were induced successfully by MAGE-A10-pulsed DCs and had cytotoxic activity which could kill lung cancer cells (Figure 5). ELISA revealed that MAGE-A10-specific CTLs produce IFN-γ which indicates that MAGE-A10-specific CTLs are cytotoxic to lung cancer cells.
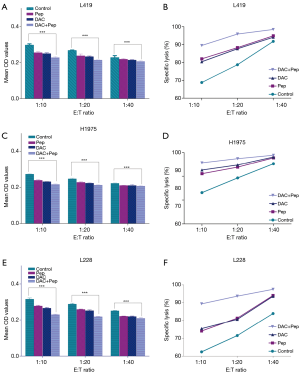
Cytotoxicity of MAGE-A10-specific CTLs was enhanced in lung cancer cells treated with DAC
To evaluate the cytotoxicity of MAGE-A10-specific CTLs on tumor cells treated with DAC, lung cancer cells treated with DAC and MAGE-A10-specific CTLs were co-cultured and then they were analyzed using CCK-8 assays and IFN-γ release assay. In the CCK-8 assays, we found that the lysis effect in the group that was treated with MAGE-A10-specific CTLs and DAC at the same time was higher than in the group that received only DAC or MAGE-A10-specific CTLs. Concomitantly, the result shows that lysis percentages increased with a rise in the E:T ratios (Figure 6).
Discussion
Lung cancer is a leading cause of cancer-related deaths worldwide. Lung cancer is usually treated with surgery, radiotherapy, and chemotherapy (16). Although there have been huge improvements in the traditional treatments, the 5-year OS for lung cancer patients has not improved significantly in the past decade (17). Therefore, there is an urgent need for novel and effective therapies.
Melanoma antigen gene family A (MAGE-A) consists of 12 closely related genes (MAGE-A1 to A12) located in the q28 region of chromosome X, that have been associated with tumor occurrence and growth and expressed in various malignant tumors (18,19). Meek et al. demonstrated that MAGE-A family is expressed in a wide range of cancers including ovary, breast, lung, and bladder (20). In addition, their expression is limited in healthy normal tissues to fetal ovary, germ cells of testis, and placenta. The restricted expression and immunogenicity of MAGE protein family makes them ideal targets for cancer immunotherapy (21,22). MAGE-A10, a subtype of the MAGE-A family, is reported to be frequently expressed in lung cancer. In our study, we found that MAGE-A10 was a strongly unfavorable prognostic indicator in human lung cancer and high expression of MAGE-A10 was significantly and negatively associated with OS [HR =1.31 Plogrank =2.1e-05, 95% CI (1.16–1.49)] and PPS [HR =1.43, Plogrank =0.0057, 95% CI (1.11–1.84)] and FP [HR =1.98, Plogrank =3.2e-12, 95% CI (1.63–2.4)].
The scientific exploitation of human immunity provides a powerful immunotherapeutic strategy to forestall various diseases. Peptides of specific antigen are proposed to be ideal candidates to be employed to provide specific immune responses as vaccines that are designed to train the human immune system to fight against a variety of diseases (23). In recent years, there is evidence that peptide vaccines can be designed with cancer antigens to activate T-cell antitumor responses, providing us with a new way to fight malignancies. The key aim of an immunotherapeutic strategy is to inhibit the ability of tumor cells to avoid recognition and elimination by the immune system in the host (24). Over the past decades, tremendous progress has been made in the understanding of how cancer evades the immune system, which in turn offers new ways to conquer malignancies (25). Multiple mechanisms employed by cancer cells, including alteration of the antigen presentation machinery or secretion of immunosuppressive factors which could induce apoptosis of lymphocytes or activate negative regulatory pathways, can induce tolerance and limit the effectiveness of the antitumor immune responses (26). In our study, although MAGE-A10 protein appears to be an attractive therapeutic target in vitro, patients with high expression of MAGE-A10 present worse clinical prognosis. This may be due to the mechanisms of long time MAGE-A10 expression related immune tolerance that operate normally in patients with lung cancer, which has been described previously.
In our study, we observed that the lysis rate of the group 1 cells was higher than that of the groups 2 and 3. This was due to the high level of expression of MAGE-A10 which enhanced the cytotoxicity of MAGE-A10-specific CTLs by improving recognition of MAGE-A10-specific CTLs in lung cancer. Therefore, our study may provide some insights on immunotherapy for those cancers that express MAGE-A10.
However, the reasons for the contradictory results of the in vitro experiments and clinical data are unclear. We hypothesize that MAGE-A10-specific CTLs are the key factors that contribute to the contradictory results. In vitro, we manually added the MAGE-A10-specific CTLs and they could recognize and kill tumor cell with MAGE-A10; whereas in vivo, the high-level expression of MAGE-A10 may inhibit the induction of MAGE-A10-specific CTLs, which results in immune tolerance of immune system leading to bad prognosis eventually. However, further experimental investigations are needed to further clarify this contradiction.
In conclusion, MAGE-A10 is an attractive therapeutic target and MAGE-A10-peptides could be exploited as vaccines in lung cancer. Thus, MAGE-A10-specific CTL can be used to improve the prognosis of lung cancer patients through killing tumor cells that overexpressMAGE-A10. This study is significant for broad-spectrum tumor immunotherapy approaches. However, further investigations are needed to determine the effectiveness of the therapy in lung cancer.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.01.10). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by institutional ethics committee (No. 2018MEC133). All procedures were in accordance with the ethical standards of the institutional ethical committee and with the 1964 Helsinki declaration and its later amendments. All applicable international, national, and/or institutional guidelines for the care were followed. All participants provided informed consent and the consent was verbal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sideras K, Bots SJ, Biermann K, et al. Tumour antigen expression in hepatocellular carcinoma in a low-endemic western area. Br J Cancer 2015;112:1911-20. [Crossref] [PubMed]
- Daudi S, Eng KH, Mhawech-Fauceglia P, et al. Expression and immune responses to MAGE antigens predict survival in epithelial ovarian cancer. PLoS One 2014;9:e104099. [Crossref] [PubMed]
- Roguljic A, Spagnoli G, Juretic A, et al. Possible predictive role of cancer/testis antigens in breast ductal carcinoma in situ. Oncol Lett 2018;16:7245-55. [PubMed]
- Mrklić I, Spagnoli GC, Juretic A, et al. Co-expression of cancer testis antigens and topoisomerase 2-alpha in triple negative breast carcinomas. Acta Histochem 2014;116:740-6. [Crossref] [PubMed]
- Border EC, Sanderson JP, Weissensteiner T, et al. Affinity-enhanced T-cell receptors for adoptive T-cell therapy targeting MAGE-A10: strategy for selection of an optimal candidate. Oncoimmunology 2018;8:e1532759. [Crossref] [PubMed]
- Ho JC, Leung CC. Management of co-existent tuberculosis and lung cancer. Lung Cancer 2018;122:83-7. [Crossref] [PubMed]
- Walter JE, Heuvelmans MA, de Bock GH, et al. Relationship between the number of new nodules and lung cancer probability in incidence screening rounds of CT lung cancer screening: The NELSON study. Lung Cancer 2018;125:103-8. [Crossref] [PubMed]
- Wang X, Leader JK, Wang R, et al. Vasculature surrounding a nodule: A novel lung cancer biomarker. Lung Cancer 2017;114:38-43. [Crossref] [PubMed]
- Lv Y, Mattson E, Bhadurihauck A, et al. Effector functions of memory CTLs can be affected by signals received during reactivation. Immunol Res 2017;65:841-52. [Crossref] [PubMed]
- Brown MC, Holl EK, Boczkowski D, et al. Cancer immunotherapy with recombinant poliovirus induces IFN-dominant activation of dendritic cells and tumor antigen-specific CTLs. Sci Transl Med 2017; [Crossref] [PubMed]
- Barranco C. Autoimmunity: CD4(+) CTLs drive IgG4-related disease. Nat Rev Rheumatol 2016;12:500. [Crossref] [PubMed]
- Maehara T, Mattoo H, Ohta M, et al. Lesional CD4+ IFN-gamma+ cytotoxic T lymphocytes in IgG4-related dacryoadenitis and sialoadenitis. Ann Rheum Dis 2017;76:377-85. [Crossref] [PubMed]
- Lauschke VM, Barragan I, Ingelman-Sundberg M. Pharmacoepigenetics and Toxicoepigenetics: Novel Mechanistic Insights and Therapeutic Opportunities. Annu Rev Pharmacol Toxicol 2018;58:161-85. [Crossref] [PubMed]
- Yu J, Qin B, Moyer AM, et al. DNA methyltransferase expression in triple-negative breast cancer predicts sensitivity to decitabine. J Clin Invest 2018;128:2376-88. [Crossref] [PubMed]
- Short NJ, Kantarjian HM, Loghavi S, et al. Treatment with a 5-day versus a 10-day schedule of decitabine in older patients with newly diagnosed acute myeloid leukaemia: a randomised phase 2 trial. Lancet Haematol 2019;6:e29-e37. [Crossref] [PubMed]
- Hamann HA, Ver HES, Carter-Harris L, et al. Multilevel Opportunities to Address Lung Cancer Stigma across the Cancer Control Continuum. J Thorac Oncol 2018;13:1062-75. [Crossref] [PubMed]
- Giroux DJ, Van Schil P, Asamura H, et al. The IASLC Lung Cancer Staging Project: A Renewed Call to Participation. J Thorac Oncol 2018;13:801-9. [Crossref] [PubMed]
- Sang M, Meng L, Sang Y, et al. Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-A family expression. Cancer Lett 2018;426:37-46. [Crossref] [PubMed]
- Fon Tacer K, Montoya MC, Oatley MJ, et al. MAGE cancer-testis antigens protect the mammalian germline under environmental stress. Sci Adv 2019;5:eaav4832.
- Meek DW, Marcar L. MAGE-A antigens as targets in tumour therapy. Cancer Lett 2012;324:126-32. [Crossref] [PubMed]
- Schooten E, Di MA, van Bergen En Henegouwen PMP, et al. MAGE-A antigens as targets for cancer immunotherapy. Cancer Treat Rev 2018;67:54-62. [Crossref] [PubMed]
- Mecklenburg I, Sienel W, Schmid S, et al. A Threshold of Systemic MAGE-A Gene Expression Predicting Survival in Resected Non-Small Cell Lung Cancer. Clin Cancer Res 2017;23:1213-9. [Crossref] [PubMed]
- Jiang H, Wang Q, Li L, et al. Turning the Old Adjuvant from Gel to Nanoparticles to Amplify CD8(+) T Cell Responses. Adv Sci (Weinh) 2017;5:1700426. [Crossref] [PubMed]
- Wildes TJ, Grippin A, Dyson KA, et al. Cross-talk between T Cells and Hematopoietic Stem Cells during Adoptive Cellular Therapy for Malignant Glioma. Clin Cancer Res 2018;24:3955-66. [Crossref] [PubMed]
- Dantzer R. Neuroimmune Interactions: From the Brain to the Immune System and Vice Versa. Physiol Rev 2018;98:477-504. [Crossref] [PubMed]
- Villani AC, Sarkizova S, Hacohen N. Systems Immunology: Learning the Rules of the Immune System. Annu Rev Immunol 2018;36:813-42. [Crossref] [PubMed]


