
The comprehensive investigation of transcription factor AP-2 alpha in lung adenocarcinoma
Introduction
Lung cancer consistently remains one of the leading causes of morbidity and mortality among patients with cancers all over the world (1,2). Despite recent advances in diagnostic and therapeutic techniques, the 5-year overall survival (OS) rate of lung cancer stilly remains poor (3). Lung adenocarcinoma (LUAD) is currently the most common sub-type of lung carcinomas (4). The early diagnosis of patients with LUAD will help patients have a relatively superior prognosis. Therefore, it is an urgent need to clarify molecular mechanisms and determine new reliable biomarkers and therapeutic targets of LUAD (5,6).
Transcription factor AP-2 alpha (TFAP2A) is an AP-2 family transcription factor, contributed to regulate cell differentiation, proliferation, and apoptosis (7,8). The protein encoded by TFAP2A gene is a transcription factor that binds the consensus sequence 5'-GCCNNNGGC-3' (7). The loss or elevation of TFAP2A expression might lead to differentiation, proliferation, and eventually to tumorigenesis, tumor progression, invasion, or metastasis (7). The dysregulation of TFAP2A has been observed in several types of cancers, such as hepatocellular carcinoma (9), breast cancer (10), gastric cancer (11), and basal-squamous bladder cancer (8). Previous studies also suggested TFAP2A was overexpressed in patients with LUAD compared with normal individuals (12,13). However, the prognostic effects of TFAP2A were not studied in the patients with LUAD. Additionally, a wider understanding of functions of TFAP2A in the LUAD required further investigation. A comprehensive study investigating the expression levels and prognostic effects of TFAP2A in the LUAD would improve the diagnosis and therapy of LUAD.
In this study, we conducted comprehensive investigation and meta-analyses to study the expression levels of TFAP2A in the patients with LUAD. Our results demonstrated TFAP2A increased in the LUAD, and the overexpression of TFAP2A was associated with worse prognosis of patients with LUAD. Furthermore, we also identified cell division cycle 6 (CDC6) and aurora kinase A (AURKA) might be two target genes of TFAP2A in the development and progress of LUAD.
Methods
GEPIA analyses
GEPIA (http://gepia.cancer-pku.cn/) provides the users opportunities to comprehensively study the differential expression, profiling plotting, patient survival, similar gene detection and dimensionality reduction analyses (14). The present study included TFAP2A into the GEPIA dataset to explore its expression values and effects on the prognosis of patients with LUAD.
Oncomine dataset analyses
The Oncomine (www.oncomine.org) dataset (15), a public cancer microarray database accessible online aiming to facilitate the discovery of cancer related genes through genome-wide expression analyses, was applied to evaluate the mRNA levels of TFAP2A between LUAD and control samples. The differences in mRNA levels of TFAP2A were performed by Students’ t-test. The threshold was established at P<10−4 and fold change (FC) >2 fold.
The Kaplan-Meier plot analyses
Prognostic values of TFAP2A in patients with LUAD were evaluated using the Kaplan-Meier plotter dataset (www.kmplot.com) (16). By far, 866 patients with LUAD have been included in the Kaplan-Meier plotter dataset. Patients were divided into high and low groups based on the median of TFAP2A expression levels. The hazard ratio (HR) with 95% confidence intervals and log rank P value were calculated to evaluate the differences between two groups.
The meta-analyses of TFAP2A in the LUAD
The Lung Cancer Explorer (LCE, http://lce.biohpc.swmed.edu/lungcancer/index.php#page-top) (17) was a lung cancer-specific database with over 6700 patients from 56 studies. The users can perform meta-analyses to study the gene expression of lung cancer patients. In the current study, we investigated the expression levels of TFAP2A in LUAD tumors vs. normal controls, and survival vs. non-survival patients.
The target genes of TFAP2A
The Animal Transcription Factor Data Base (Animal TFDB3.0, http://bioinfo.life.hust.edu.cn/AnimalTFDB/) is a resource aiming to provide the most comprehensive and accurate information for transcription factors (TFs) and cofactors (18). In the current study, we studied the targeted genes of TFAP2A in Animal TFDB 3.0.
The GO and KEGG pathway analyses
To obtain an insight into the target genes of TFAP2A at the functional level, Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were accomplished using the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/). FDR <0.05 was considered to indicate a statistically significant difference.
Results
The different transcriptional levels of TFAP2A in patients with LUAD
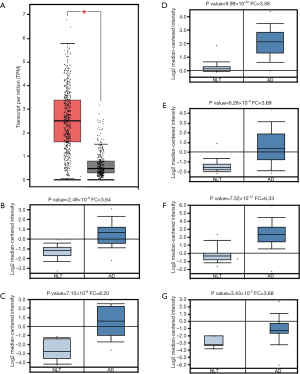
To study the expression levels of TFAP2A in the patients with LUAD, we investigated the transcription levels of TFAP2A in the GEPIA and Oncomine databases. In the GEPIA database, the expression levels of TFAP2A were studied in 483 patients with LUAD and 347 normal individuals. The results indicated the transcriptional expression levels of TFAP2A were significantly elevated in the patients with LUAD (P<0.01) (Figure 1A). Up to now, six studies had indicated TFAP2A was significantly elevated in patients with LUAD in the Oncomine database. The study by Su et al. (19) on LUAD suggested the TFAP2A mRNA levels increased 3.64-fold compared with normal samples (P=2.48×10−8) (Figure 1B). The study on LUAD conducted by Stearman et al. (20) revealed the transcription levels of TFAP2A elevated 8.20-fold compared with normal samples (P=7.10×10−8) (Figure 1C). The study conducted by Selamat et al. (21) demonstrated the levels of TFAP2A mRNA were 3.58-fold elevated in LUAD samples compared with normal samples (P=9.99×10−20) (Figure 1D). In Hou et al.’s study (22), the levels of TFAP2A mRNA were elevated by 3.69-fold compared with normal lung samples (P=6.28×10−9) (Figure 1E). Compared with normal lung samples, TFAP2A levels increased 6.33-fold in Landi et al.’s LUAD group (P=7.52×10−12) (Figure 1E) (23). Compared with normal lung tissues, the TFAP2A mRNA levels increased 3.68-fold in Garber et al.’s LUAD group (P=3.43×10−5) (Figure 1F) (24). Therefore, the overexpression of TFAP2A in the patients with LUAD was confirmed.
The prognostic values of TFAP2A in patients with LUAD
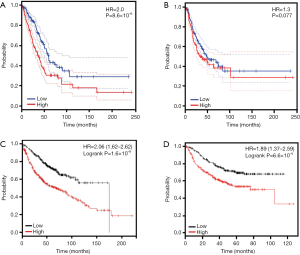
To study the prognostic effects of TFAP2A in patients with LUAD, we studied the association between transcription levels of TFAP2A and survival time of patients with LUAD. In the GEPIA dataset, our results indicated patients with high-TFAP2A expression exhibited shorter OS periods than those with low-TFAP2A expression (HR=2.0, P=8.6×10−6) (Figure 2A). Meanwhile, our study demonstrated LUAD patients with high-TFAP2A expression had shorter progression-free-survival (PFS) periods than those with low-TFAP2A (HR=1.3, P=0.077) (Figure 2B). Additionally, we validated the prognostic effects of the TFAP2A mRNA in the patients with LUAD using the Kaplan-Meier plotter dataset. Kaplan-Meier analyses revealed patients with LUAD having higher TFAP2A levels exhibited shorter OS periods than those with lower TFAP2A levels [HR=2.06 (1.62–2.62), P=1.6×10−9] (Figure 2C). Meanwhile, Kaplan-Meier analyses also indicated patients with higher TFAP2A suggested shorter PFS periods than those with lower TFAP2A levels [HR=1.89 (1.37–2.59), P=6.6×10−5] (Figure 2D).
The meta-analyses of TFAP2A in patients with LUAD
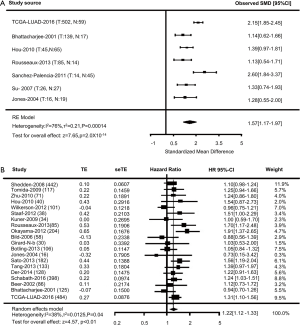
To further validate our results, meta-analyses of TFAP2A in patients with LUAD were performed in the LCE database. A comparative analysis suggested 7 datasets were conducted to study the expression levels of TFAP2A between patients with LUAD (n=827) and normal individuals (n=246). As shown in Figure 3A, significant evidence of heterogeneity was observed (I2=76%, P=1.40×10−4), and a random-effects model was conducted to calculate the Standardized Mean Difference (SMD) among all 7 datasets. The overall SMD for 7 studies was 1.57 (1.17–1.97) (P=2.0×10−14) (Figure 3A). Therefore, our meta-analyses results indicated patients with LUAD have higher TFAP2A levels than normal individuals. Meanwhile, we evaluated the expression levels of TFAP2A between survival and non-survival patients with LUAD. Finally, 20 studies with 2,878 patients with LUAD were included for meta-analyses. As shown in Figure 3B, significant evidence of heterogeneity was detected in all studies (I2=38%, P=0.04), and a random-effects model was carried out to calculate the HR. Overall HR for 20 studies was 1.22 (1.12–1.33) (P<0.01) (Figure 3B). Therefore, we demonstrated the levels of TFAP2A up-regulated in patients with LUAD, and the expression levels of TFAP2A might affect the prognosis of patients with LUAD.
The target genes of TFAP2A in the patients with LUAD
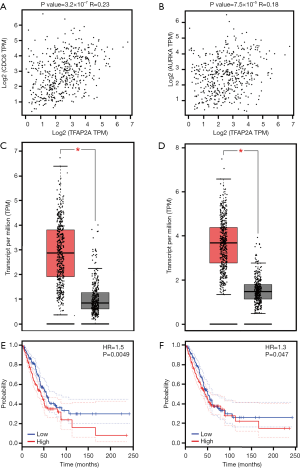
To identify the potential target genes of TFAP2A, we investigated the TFAP2A in the Animal TFDB3.0 database. As shown in Figure 4, 63 target genes of TFAP2A were identified. Then, GO analyses indicated those genes were related to positive regulation of metabolic process, positive regulation of macromolecule metabolic process and positive regulation of nitrogen compound metabolic process (Table 1). KEGG analyses suggested those genes were associated with pathways in cancer, mitophagy and Wnt signaling pathway (Table 2). Then, we uploaded those genes into GEPIA dataset to study the correlations with TFAP2A. Our results found only cell division cycle 6 (CDC6) (R=0.23, P=3.2×10−7) (Figure 5A) and aurora kinase A (AURKA) (R=0.18, P=7.5×10−5) (Figure 5B) were significantly correlated with the expression levels of TFAP2A in patients with LUAD. Meanwhile, CDC6 and AURKA were elevated in the patients with LUAD (P<0.01) (Figure 5C,D). Furthermore, CDC6 and AURKA were significantly associated with the prognosis of patients with LUAD (HR=1.5, P=0.0049 and HR=1.3, P=0.047, respectively) (Figure 5E,F). Therefore, we considered CDC6 and AURKA might be two potential targets of TFAP2A in the patients with LUAD.
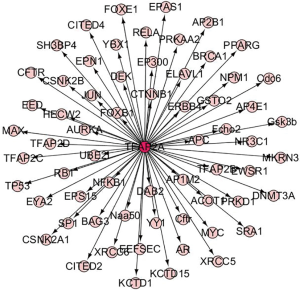
Table 1
| GO term | Term description | Counts | FDR |
|---|---|---|---|
| Biological process | |||
| GO:0009893 | Positive regulation of metabolic process | 45 | 4.23×10−18 |
| GO:0010604 | Positive regulation of macromolecule metabolic process | 44 | 4.23×10−18 |
| GO:0051173 | Positive regulation of nitrogen compound metabolic process | 42 | 3.28×10−17 |
| GO:0031325 | Positive regulation of cellular metabolic process | 42 | 1.04×10−16 |
| GO:2000112 | Regulation of cellular macromolecule biosynthetic process | 46 | 4.87×10−16 |
| Molecular function | |||
| GO:0044212 | Transcription regulatory region DNA binding | 24 | 1.76×10−16 |
| GO:0043565 | Sequence-specific DNA binding | 24 | 9.57×10−13 |
| GO:0008134 | Transcription factor binding | 19 | 4.80×10−12 |
| GO:0003690 | Double-stranded DNA binding | 21 | 6.61×10−12 |
| GO:0000987 | Proximal promoter sequence-specific DNA binding | 17 | 1.01×10−11 |
| Cellular component | |||
| GO:0005654 | Nucleoplasm | 41 | 3.71×10−14 |
| GO:0031981 | Nuclear lumen | 43 | 8.59×10−14 |
| GO:0044454 | Nuclear chromosome part | 19 | 8.59×10−14 |
| GO:0005634 | Nucleus | 52 | 9.26×10−13 |
| GO:0044427 | Chromosomal part | 21 | 3.21×10−12 |
GO, gene ontology.
Table 2
| KEGG term | Term description | Counts | FDR |
|---|---|---|---|
| hsa05200 | Pathways in cancer | 16 | 1.16×10−9 |
| hsa04137 | Mitophagy | 7 | 7.28×10−8 |
| hsa04310 | Wnt signaling pathway | 9 | 7.28×10−8 |
| hsa05169 | Epstein-Barr virus infection | 10 | 7.28×10−8 |
| hsa05215 | Prostate cancer | 8 | 7.28×10−8 |
KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
The comprehensive investigation of TFAP2A in the patients with LUAD would help to develop TFAP2A-targeted therapy. In the present study, we evaluated the dysregulation and the prognostic effects of TFAP2A in patients with LUAD using bioinformatics methods from the public databases. Our study found TFAP2A upregulated in patients with LUAD, and the increasing TFAP2A was associated with shorter survival time of patients with LUAD. Finally, CDC6 and AURKA were considered as two potential targeted genes of TFAP2A. Our study also demonstrated CDC6 and AURKA might serve as feature genes closely related to the development and prognosis of patients with LUAD.
Previous studies demonstrated TFAP2A could exhibit either oncogenic or suppressive characteristics in various tumor models (7). As an oncogene, TFAP2A modulated the expression of multiple target genes involved in the development and progression of human cancers. Shi et al.’s (25) study suggested TFAP2A highly expressed in various nasopharyngeal carcinoma cell lines and tumor tissue specimens. A positive correlation of TFAP2A overexpression with advanced tumor stage, local invasion, clinical progression, and poor prognosis of patients with nasopharyngeal carcinomas were also observed. Yamashita et al.’s (8) study suggested TFAP2A are significantly overexpressed in basal-squamous bladder cancer, and TFAP2A overexpression is associated with the presence of lymph node metastasis and distant recurrence. In our current study, we indicated TFAP2A upregulated in patients with LUAD, which was consistent with Yuanhua et al.’s and Pu et al.’s studies (12,13). Moreover, our study also demonstrated higher TFAP2A was associated with shorter survival time of patients with LUAD. Therefore, we considered TFAP2A might be an oncogene in the development and progression of LUAD. However, the mechanisms how TFAP2A affected the development of LUAD were poorly studied. The Yuanhua et al.’s (12) study indicated TFAP2A induced the expression of KRT16 promoting the tumorigenicity of LUAD through affecting the epithelial-mesenchymal transition (EMT). In Pu et al.’s study (13), the cell apoptosis and cell cycle of non-small cell lung cancer cell were induced by MiR-1254 via inhibited HO-1 transcription by suppressing the TFAP2A. These results suggested TFAP2A might influence a wide range of physiological or pathological processes by regulating different pathways and interacting with specific molecules (7). Therefore, we speculated that TFAP2A should be explored as a therapeutic target of patients with LUAD.
To better understand the role of TFAP2A in the LUAD, we investigated the potential target genes of TFAP2A. In our study, only the CDC6 and AURKA were considered to affect the development and progress of LUAD. The CDC6 could regulate DNA replication licensing, tumorigenesis, and prognosis in lung cancer (26). Li et al.’s reports suggested CDC6 might serve as a potential prognostic marker of LUAD (27). He et al.’s study also indicated CDC6 might link LUAD to smoking history, and CDC6 will be useful for future clinical studies (28). Meanwhile, previous studies demonstrated AURKA was potential biomarker for predicting poor prognosis of LUAD (27,29). AURKA played important roles in the proliferation of human LUAD cells, which suggested AURKA could be a promising tool for the therapy of LUAD (30). Meta-analyses also identified AURKA was the most obvious class of hub nodes (31). Therefore, we considered TFAP2A might contribute to the development of LUAD via affecting the expression of AURKA and CDC6. However, deeply associations among TFAP2A, AURKA and CDC6 were unknown, further studies were warranted to investigate the mechanisms.
In the current study, dysregulation of TFAP2A was investigated in the patients with LUAD. Up-regulation of TFAP2A and its clinical significance were validated in various large online databases. However, some limitations also were obvious. First, lung cancer had several subgroups, we only investigate the TFAP2A expression levels in patients with LUAD, further investigations of TFAP2A in other lung cancer subgroups were necessary. Second, although we evaluated the overexpression of TFAP2A mRNA in various public databases, the protein levels of TFAP2A were not studied in our research. Finally, the mechanisms of TFAP2A in the development of LUAD needed to be studied in the future study.
Conclusions
In conclusion, our study had performed comprehensive analyses to study the TFAP2A in the progress of LUAD. Elevated TFAP2A was detected and validated in the patients with LUAD. The higher TFAP2A were associated with the worse prognosis of patients with LUAD. Moreover, CDC6 and AURKA were considered as two target genes of TFAP2A. However, further molecular biological experiments were required to investigate mechanisms of TFAP2A in LUAD.
Acknowledgments
We acknowledge Dr. Dong Jiang and Yang Zhang for data analysis and manuscript writing.
Funding: None.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.01.58). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 2019;144:1941-53. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Han X, Tan Q, Yang S, et al. Comprehensive Profiling of Gene Copy Number Alterations Predicts Patient Prognosis in Resected Stages I-III Lung Adenocarcinoma. Front Oncol 2019;9:556. [Crossref] [PubMed]
- Fernandes MGO, Jacob M, Martins N, et al. Targeted Gene Next-Generation Sequencing Panel in Patients with Advanced Lung Adenocarcinoma: Paving the Way for Clinical Implementation. Cancers (Basel) 2019;11: [Crossref] [PubMed]
- Scafoglio CR, Villegas B, Abdelhady G, et al. Sodium-glucose transporter 2 is a diagnostic and therapeutic target for early-stage lung adenocarcinoma. Sci Transl Med 2018;10: [Crossref] [PubMed]
- Dong X, Zhang R, He J, et al. Trans-omics biomarker model improves prognostic prediction accuracy for early-stage lung adenocarcinoma. Aging (Albany NY) 2019;11:6312-35. [Crossref] [PubMed]
- Kołat D, Kaluzinska Z, Bednarek AK, et al. The biological characteristics of transcription factors AP-2alpha and AP-2gamma and their importance in various types of cancers. Biosci Rep 2019;39. [PubMed]
- Yamashita H, Kawasawa YI, Shuman L, et al. Repression of transcription factor AP-2 alpha by PPARgamma reveals a novel transcriptional circuit in basal-squamous bladder cancer. Oncogenesis 2019;8:69. [Crossref] [PubMed]
- Huang W, Chen C, Liang Z, et al. AP-2alpha inhibits hepatocellular carcinoma cell growth and migration. Int J Oncol 2016;48:1125-34. [Crossref] [PubMed]
- Xu J, Zheng J, Wang J, et al. miR-876-5p suppresses breast cancer progression through targeting TFAP2A. Exp Ther Med 2019;18:1458-64. [PubMed]
- Cao L, Wang S, Zhang Y, et al. Zinc-finger protein 471 suppresses gastric cancer through transcriptionally repressing downstream oncogenic PLS3 and TFAP2A. Oncogene 2018;37:3601-16. [Crossref] [PubMed]
- Yuanhua L, Pudong Q, Wei Z, et al. TFAP2A Induced KRT16 as an Oncogene in Lung Adenocarcinoma via EMT. Int J Biol Sci 2019;15:1419-28. [Crossref] [PubMed]
- Pu M, Li C, Qi X, et al. MiR-1254 suppresses HO-1 expression through seed region-dependent silencing and non-seed interaction with TFAP2A transcript to attenuate NSCLC growth. PLoS Genet 2017;13:e1006896. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Nagy A, Lanczky A, Menyhart O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. [Crossref] [PubMed]
- Cai L, Lin S, Girard L, et al. LCE: an open web portal to explore gene expression and clinical associations in lung cancer. Oncogene 2019;38:2551-64. [Crossref] [PubMed]
- Hu H, Miao YR, Jia LH, et al. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res 2019;47:D33-8. [Crossref] [PubMed]
- Su LJ, Chang CW, Wu YC, et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics 2007;8:140. [Crossref] [PubMed]
- Stearman RS, Dwyer-Nield L, Zerbe L, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol 2005;167:1763-75. [Crossref] [PubMed]
- Selamat SA, Chung BS, Girard L, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res 2012;22:1197-211. [Crossref] [PubMed]
- Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One 2010;5:e10312. [Crossref] [PubMed]
- Landi MT, Dracheva T, Rotunno M, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One 2008;3:e1651. [Crossref] [PubMed]
- Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A 2001;98:13784-9. [Crossref] [PubMed]
- Shi D, Xie F, Zhang Y, et al. TFAP2A regulates nasopharyngeal carcinoma growth and survival by targeting HIF-1alpha signaling pathway. Cancer Prev Res (Phila) 2014;7:266-77. [Crossref] [PubMed]
- Zhang X, Xiao D, Wang Z, et al. MicroRNA-26a/b regulate DNA replication licensing, tumorigenesis, and prognosis by targeting CDC6 in lung cancer. Mol Cancer Res 2014;12:1535-46. [Crossref] [PubMed]
- Li S, Xuan Y, Gao B, et al. Identification of an eight-gene prognostic signature for lung adenocarcinoma. Cancer Manag Res 2018;10:3383-92. [Crossref] [PubMed]
- He X, Zhang C, Shi C, et al. Meta-analysis of mRNA expression profiles to identify differentially expressed genes in lung adenocarcinoma tissue from smokers and non-smokers. Oncol Rep 2018;39:929-38. [PubMed]
- Zhang MY, Liu XX, Li H, et al. Elevated mRNA Levels of AURKA, CDC20 and TPX2 are associated with poor prognosis of smoking related lung adenocarcinoma using bioinformatics analysis. Int J Med Sci 2018;15:1676-85. [Crossref] [PubMed]
- Zhong N, Shi S, Wang H, et al. Silencing Aurora-A with siRNA inhibits cell proliferation in human lung adenocarcinoma cells. Int J Oncol 2016;49:1028-38. [Crossref] [PubMed]
- Selvaraj G, Kaliamurthi S, Kaushik AC, et al. Identification of target gene and prognostic evaluation for lung adenocarcinoma using gene expression meta-analysis, network analysis and neural network algorithms. J Biomed Inform 2018;86:120-34. [Crossref] [PubMed]


