Identification of three miRNAs signature as a prognostic biomarker in breast cancer using bioinformatics analysis
Introduction
Breast cancer is the most common neoplasm in females and the second leading reason of cancer death in females. It is estimated that approximately 62,930 cases of cancer in situ of the female mammary gland were newly diagnosed in 2019 (1). Although progress has been made in neoadjuvant chemotherapy and surgical techniques, some breast cancer patients still have poor prognosis, especially for human epidermal growth factor receptor-2 (HER-2) positive breast cancer or triple negative breast cancer (TNBC) patients (2). Therefore, uncovering the pathogenesis and identifying new biomarkers are urgently needed for early diagnosis, judge prognosis, and direct treatment for breast cancer.
MicroRNAs (miRNAs) are small noncoding RNAs that inhibit the expression of target genes by targeting the 3'UTR of mRNA (3). Accumulating evidences showed that miRNAs are unusually expressed in many different kinds of neoplasms and play important roles in tumorigenesis and development (4). It has been demonstrated that miRNAs modulated a variety of oncogenic processes, including cell viability (5) apoptosis (6), autophagy (7), migration, invasion (8), and cell maturation (9). Therefore, identification of tumor-specific miRNAs will have important implications for predicting prognosis in breast cancer patients.
In the current study, we identified differentially expression miRNAs (DEmiRNAs) between normal breast tissues and breast cancer tissues by analyzing the sequencing data of miRNAs downloaded from The Cancer Genome Atlas (TCGA) database. Moreover, we found that three miRNA signatures may validly predict the prognosis of patients. Additionally, we evaluated the function and signaling pathway of predicting gene with Enrichr database, which might provide further understand the molecular mechanism of breast cancer.
Methods
Data acquisition and characteristics
We downloaded clinical information and raw sequencing data from the official TCGA website (https://cancergenome.nih.gov/). We selected samples with miRNA sequencing data and relevant clinical prognostic data for subsequent analysis. Finally, our study included 1,166 samples, including 104 normal samples and 1,062 breast cancer samples. Clinical information including age at diagnosis, gender, clinical stage, lymph-node status, T stage, metastasis, progesterone receptor (PR) status, estrogen receptor (ER) status, and Her2 status, and is shown in Table 1.
Table 1
| Variables | Case, n (%) |
|---|---|
| Age at diagnosis (years) | |
| ≥60 | 494 (46.5) |
| <60 | 568 (53.5) |
| Gender | |
| Male | 12 (1.1) |
| Female | 1,050 (98.9) |
| Metastasis | |
| M0 | 879 (82.8) |
| M1 | 21 (2.0) |
| MX | 162 (15.3) |
| Lymph-node status | |
| N0 | 498 (46.9) |
| N1−3 | 544 (51.2) |
| NX | 20 (1.9) |
| Clinical stage | |
| I+II | 779 (73.4) |
| III+IV | 261 (24.6) |
| NA | 22 (2.1) |
| T stage | |
| T1+T2 | 892 (84.0) |
| T3+T4 | 167 (15.7) |
| TX | 3 (0.3) |
| ER status | |
| Positive | 782 (73.6) |
| Negative | 232 (21.8) |
| NA | 48 (4.5) |
| PR status | |
| Positive | 680 (64.0) |
| Negative | 331 (31.2) |
| NA | 51 (4.8) |
| Her2 status | |
| Positive | 173 (16.3) |
| Negative | 748 (70.4) |
| NA | 141 (13.3) |
NA, non-available.
Raw-count miRNA sequencing data were conducted with Edge-R to screen DEmiRNAs between normal tissues and breast cancer tissues. Log2|fold changes (FCs)| >2.0 and FDR values <0.05 were set as cut-off criteria for identifying DEmiRNAs.
Correlation analysis between DEmiRNAs and prognosis of breast cancer
The prognostic value of each DEmiRNA were estimated by log-rank test and Kaplan-Meier method. We first identified miRNAs correlated with overall survival (OS) and then performed binary logistic regression analysis. Then, we constructed prognostic miRNA signature, and it can count risk score for each patient. Based on the median risk score of miRNA, patients with breast cancer were divided into high- and low-risk groups. Next, we assessed the difference in survival between the two groups by using Kaplan-Meier method.
Target gene prediction and functional enrichment analysis
Three miRNAs-targeted mRNAs were predicted by using three online analytical softwares: miRDB, TargetScan, and miRDIP. In order to further improve the credibility of bioinformatics analysis, we then used Venn diagram to screen the overlapping target genes. Finally, we evaluated the function and signaling pathway of predicting gene with Enrichr bioinformatics tool (http://amp.pharm.mssm.edu/Enrichr/).
Statistical analysis
The relationship between patient clinicopathological parameters and miRNA expression level was assessed by using t-test and Chi-square test, as appropriate. The correlation between three-miRNA signature and patients’ survival was assessed by Kaplan-Meier method. The prognostic value of three miRNAs were analyzed by Cox analysis. All statistical analyses were performed using R (v.3.6.0) software and SPSS v.24.0 software (IBM Corp.).
Results
Identification of DEmiRNAs in breast cancer
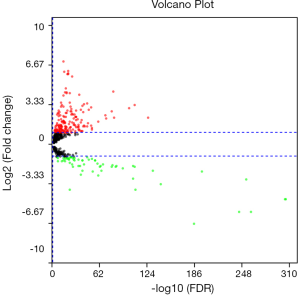
Using FDR <0.05 and |log2FC| ≥2 as the cut-off criteria. We identified 84 differentially expressed miRNAs (DEmiRNAs), among them, 17 were downregulated and 67 were upregulated (Table 2). The relationships between log2FC and FDR of each DEmiRNA were shown by the volcano plot (Figure 1).
Table 2
| miRNA | logFC | P value | FDR |
|---|---|---|---|
| Upregulated | |||
| hsa-miR-122 | 7.024277 | 8.59E-15 | 2.39E-14 |
| hsa-miR-1269a | 6.219576 | 2.25E-21 | 9.05E-21 |
| hsa-miR-1269b | 6.175296 | 1.14E-16 | 3.58E-16 |
| hsa-miR-767 | 6.047708 | 1.42E-21 | 5.80E-21 |
| hsa-miR-105-2 | 5.999491 | 2.13E-20 | 8.38E-20 |
| hsa-miR-105-1 | 5.985519 | 1.53E-21 | 6.23E-21 |
| hsa-miR-449a | 5.736712 | 3.68E-27 | 1.98E-26 |
| hsa-miR-3156-1 | 4.752563 | 2.21E-18 | 7.79E-18 |
| hsa-miR-592 | 4.52489 | 9.63E-81 | 2.39E-79 |
| hsa-miR-449c | 4.442171 | 7.59E-19 | 2.70E-18 |
| hsa-miR-3156-3 | 4.419179 | 1.92E-17 | 6.37E-17 |
| hsa-miR-184 | 4.393167 | 5.11E-24 | 2.35E-23 |
| hsa-miR-449b | 4.357878 | 3.22E-19 | 1.17E-18 |
| hsa-miR-4724 | 4.221601 | 9.71E-36 | 7.91E-35 |
| hsa-miR-3156-2 | 4.198612 | 7.71E-15 | 2.17E-14 |
| hsa-miR-522 | 3.581905 | 4.43E-12 | 1.06E-11 |
| hsa-miR-4501 | 3.540763 | 1.41E-23 | 6.30E-23 |
| hsa-miR-190b | 3.524598 | 2.84E-47 | 3.45E-46 |
| hsa-miR-196a-1 | 3.428353 | 1.03E-42 | 1.13E-41 |
| hsa-miR-96 | 3.397427 | 9.58E-109 | 3.64E-107 |
| hsa-miR-1251 | 3.344711 | 3.58E-13 | 9.18E-13 |
| hsa-miR-196a-2 | 3.284369 | 5.45E-41 | 5.54E-40 |
| hsa-miR-4652 | 3.264641 | 9.98E-15 | 2.75E-14 |
| hsa-miR-187 | 3.260218 | 2.57E-24 | 1.22E-23 |
| hsa-miR-519a-1 | 3.230734 | 1.78E-12 | 4.43E-12 |
| hsa-miR-210 | 3.191708 | 3.98E-52 | 5.40E-51 |
| hsa-miR-183 | 3.073046 | 4.16E-102 | 1.32E-100 |
| hsa-miR-7-3 | 3.013492 | 2.96E-25 | 1.47E-24 |
| hsa-miR-301b | 2.993863 | 7.22E-39 | 6.53E-38 |
| hsa-miR-7705 | 2.975935 | 1.72E-48 | 2.18E-47 |
| Downregulated | |||
| hsa-miR-133b | −6.64828 | 1.77E-187 | 1.12E-185 |
| hsa-miR-1-2 | −5.66393 | 3.31E-261 | 3.78E-259 |
| hsa-miR-1-1 | −5.64985 | 2.02E-246 | 1.65E-244 |
| hsa-miR-486-1 | −4.56451 | 2.99E-307 | 5.67E-305 |
| hsa-miR-486-2 | −4.56384 | 1.12E-306 | 1.59E-304 |
| hsa-miR-206 | −3.78198 | 4.67E-24 | 2.16E-23 |
| hsa-miR-4732 | −3.7358 | 3.35E-110 | 1.47E-108 |
| hsa-miR-451a | −3.29369 | 5.93E-144 | 3.38E-142 |
| hsa-miR-139 | −2.90964 | 1.04E-255 | 9.84E-254 |
| hsa-miR-144 | −2.86216 | 8.29E-107 | 2.78E-105 |
| hsa-miR-204 | −2.5508 | 1.09E-61 | 1.88E-60 |
| hsa-miR-6715a | −2.5086 | 3.15E-38 | 2.76E-37 |
| hsa-miR-145 | −2.24857 | 9.89E-198 | 7.05E-196 |
| hsa-miR-1258 | −2.21095 | 2.29E-54 | 3.43E-53 |
| hsa-miR-378a | −2.17398 | 3.13E-133 | 1.62E-131 |
| hsa-miR-5683 | −2.16241 | 8.46E-41 | 8.31E-40 |
| hsa-miR-551b | −2.14579 | 5.10E-66 | 9.69E-65 |
DEmiRNAs, expressed miRNAs; miRNA, MicroRNAs.
Screening for miRNAs correlated with OS
To further understand the correlation between differently expressed miRNAs and OS, we performed Kaplan-Meier analysis. The results indicated that miR-105-1, miR-301b and miR-1258 were significantly related with breast cancer patients’ OS (Figure 2). As shown in Table 3, the correlation between the expression of three kinds of miRNA and clinicopathological parameters was assessed in breast cancer patients. Our research revealed that miR-105-1 was obviously correlated with ER and PR; miR-301b was correlated with Age, Metastasis, PR, ER, and HER-2. miR-1258 was correlated with age, clinical stage, PR, and ER.
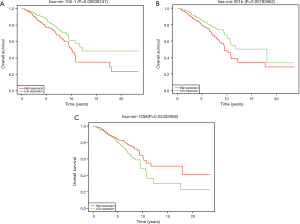
Table 3
| Variables | Case | miR-105-1 | miR-301b | miR-1258 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High | Low | P | High | Low | P | High | Low | P | ||||
| Age at diagnosis | 0.977 | 0.014 | 0.026 | |||||||||
| ≥60 | 494 | 250 | 244 | 223 | 271 | 234 | 260 | |||||
| <60 | 568 | 281 | 287 | 308 | 260 | 297 | 271 | |||||
| Gender | 0.082 | 0.082 | 0.082 | |||||||||
| Male | 12 | 9 | 3 | 9 | 3 | 3 | 9 | |||||
| Female | 1050 | 522 | 528 | 522 | 528 | 528 | 522 | |||||
| Metastasis | 0.125 | 0.023 | 0.135 | |||||||||
| M0 | 879 | 437 | 442 | 449 | 430 | 441 | 438 | |||||
| M1 | 21 | 14 | 7 | 16 | 5 | 14 | 7 | |||||
| Lymph-node status | 0.457 | 0.590 | 0.860 | |||||||||
| N0 | 498 | 255 | 243 | 250 | 248 | 227 | 271 | |||||
| N1-3 | 544 | 266 | 278 | 264 | 280 | 245 | 299 | |||||
| Clinical stage | 0.397 | 0.153 | 0.002 | |||||||||
| I+II | 779 | 380 | 399 | 395 | 384 | 369 | 410 | |||||
| III+IV | 261 | 140 | 121 | 119 | 142 | 152 | 109 | |||||
| T stage | 0.546 | 0.361 | 0.564 | |||||||||
| T1+T2 | 892 | 442 | 450 | 451 | 441 | 443 | 449 | |||||
| T3+T4 | 167 | 87 | 80 | 78 | 89 | 87 | 80 | |||||
| ER status | <0.001 | <0.001 | <0.001 | |||||||||
| Positive | 782 | 335 | 447 | 323 | 459 | 419 | 363 | |||||
| Negative | 232 | 169 | 63 | 180 | 52 | 89 | 143 | |||||
| PR status | <0.001 | <0.001 | <0.001 | |||||||||
| Positive | 680 | 276 | 404 | 277 | 403 | 373 | 307 | |||||
| Negative | 331 | 226 | 105 | 225 | 106 | 133 | 198 | |||||
| HER-2 status | 0.074 | 0.003 | 0.444 | |||||||||
| Positive | 173 | 97 | 76 | 103 | 70 | 83 | 90 | |||||
| Negative | 748 | 363 | 385 | 352 | 396 | 383 | 365 | |||||
Prognostic value of three-miRNA signature
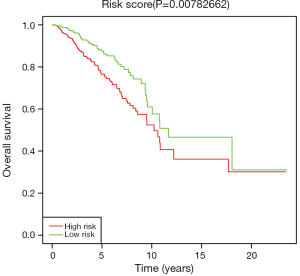
We used Kaplan-Meier method to assess the survival performance of three-miRNA signature, and estimated the risk score for every breast cancer patient. According to the median risk score, we divided breast cancer patients into low and high risk groups. As shown in Figure 3, the OS rate of breast cancer patients with high-risk scores was lower than that of patients with low-risk scores (P=0.0078).
Then, we performed univariate Cox regression analysis for each clinicopathological factor. As shown in Table 4, age (HR =1.031, P<0.001), Metastasis (HR =6.417, P<0.001), lymph node metastasis (HR =2.314, P<0.001), Clinical stage (HR =2.702, P<0.001), ER (HR =0.683, P=0.043), and three-miRNA signature (HR =1.777, P=0.001) were significant related with survival in breast cancer patients. Subsequently, we carried out the multivariate Cox regression analysis of the factors, and observed that three-miRNA signature (HR =1.574, P=0.014) was an independent prognostic factor for poor prognosis of breast malignant tumor patient (Table 4).
Table 4
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Age (≥60 vs. <60) | 1.031 (1.018–1.044) | <0.001 | 1.031 (1.018–1.044) | <0.001 | |
| Gender (male vs. female) | 0.833 (0.116–5.965) | 0.855 | |||
| Metastasis (M1 vs. M0) | 6.417 (3.795–10.851) | <0.001 | 3.540 (1.962–6.385) | <0.001 | |
| Lymph-node status (N1-3 vs. N0) | 2.314 (1.608–3.329) | <0.001 | 1.709 (1.099–2.658) | 0.017 | |
| Clinical stage (III+IV vs. I+II) | 2.702 (1.930–3.781) | <0.001 | 1.917 (1.233–2.979) | 0.004 | |
| T stage (T3+T4 vs. T1+T2) | 1.219 (0.807–1.842) | 0.346 | |||
| ER status (positive vs. negative) | 0.683 (0.472–0.988) | 0.043 | 0.579 (0.390–0.861) | 0.007 | |
| PR status (positive vs. negative) | 0.727 (0.516–1.023) | 0.067 | |||
| HER-2 status (positive vs. negative) | 1.150 (0.697–1.896) | 0.585 | |||
| Three-miRNA signature (high risk vs. low risk) | 1.777 (1.271–2.484) | 0.001 | 1.574 (1.098–2.257) | 0.014 | |
Pathway enrichment analysis of predicted target genes
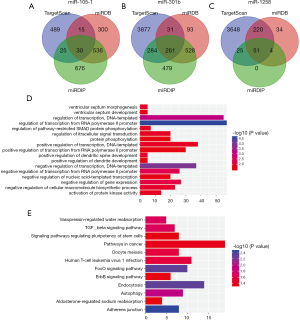
We used miRDB, TargetScan and miRDIP online analysis softwares to predict target genes for three miRNAs. The overlapping genes of miR-150-1, miR-301b and miR-1258 were identified as 30, 261 and 51, respectively (Figure 4). To further elucidate the biological roles of differently expressed miRNAs, we conducted Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) analysis to indicate the molecular function of predicted genes. Biological process was obviously enriched in signal transduction, regulation of transcription, activation of protein kinase activity. Additionally, KEGG signaling pathways were mainly concentrated in adherens junction, autophagy, and TGF-beta signaling pathway.
Discussion
Breast cancer is the second leading reason of cancer death in females (10). Survival rate of breast cancer patients will be significantly improved if cancer behavior can be reliably predicted in the initial diagnosis of the disease. Accordingly, it is extremely significant to explore the molecular mechanisms of breast cancer development and to identify the specific and sensitive biomarkers.
In this study, we identified 84 differentially expressed miRNAs (DEmiRNAs). Among them, 17 were downregulated and 67 were upregulated, and three of them were related with the OS in breast cancer patients. It has been indicated that combinations of differently miRNAs might be more specific and sensitive than single miRNA biomarkers (11). In our study, we constructed a three miRNAs signature (miR-105-1, miR-301b and miR-1258) and found that this signature was an independent prognostic indicator for breast cancer patients. Furthermore, we used three online bioinformatics tools to predict target genes for these three miRNAs, and evaluated cellular functions and signal pathway of those three miRNAs by GO and KEGG pathway analysis. In the last decade, some evidences have indicated that several miRNAs play key roles in various pathological activities, such as cell growth, migration and invasion, differentiation, cell cycle arrest, apoptosis, and autophagy (12-16). It has been suggested that miRNAs could act as novel prognostic biomarkers and therapeutic targets (17,18). Nevertheless, previous researches were based on various detection platforms, and the number of miRNAs was relatively limited. In the current study, we analyzed the sequencing data of miRNA and found that upregulation of miR-105-1 and miR-301b expression and downregulation of miR-1258 expression were clearly correlated with the prognosis of the patients with breast cancer. Previous studies indicated that exosomal miR-105 can promote cancer cell migration and distant metastasis by targeting zona occludens protein 1 (ZO-1) (19). Jin et al. showed that miR-105 promotes epithelial mesenchymal transition (EMT) via upregulation of myeloid cell leukemia-1 (MCL-1) in non-small cell lung cancer (NSCLC) (20). miR-105 was also showed to activate Wnt/β-catenin signaling by targeting secreted frizzed related protein (SFRP1), and promote chemoresistance, stemness, and metastasis in TNBC. In addition, it has been shown that the expression of miR-301 was up-regulated in some cancers, including gastric, pancreatic, hepatocellular, lung, and breast cancers (21-25). Overexpression of miR-301b can increases autophagy, viability and radioresistance in prostate cancer (26,27). Song et al. demonstrated that miR-301b can play a carcinogenic role in TNBC by targeting cylindromatosis (CYLD) (28). miR-1258 was reported to suppress breast cancer brain metastasis by inhibiting heparanase (29). Other studies also indicated miR-1258 may function in oral squamous cell carcinoma, colorectal cancer, NSCLC, and osteosarcoma (30-33). Our results showed that miR-105-1 was obviously correlated with ER and PR; miR-301b was associated with age, metastasis, ER, PR, and HER-2; miR-1258 was associated with age, clinical stage, ER, and PR, indicating that these three-miRNAs were participated in the progression of breast cancer.
In our present study, we observed that miR-105-1, miR-301b and miR-1258 were significantly correlated with OS in patients with breast cancer. We then established a three-miRNA signature, and found that this signature was a potential independent prognostic factor for breast cancer patients.
In order to further explore the biology functions of these three miRNAs, we conducted KEGG pathway and Gene Ontology analysis to indicate the molecular function of predicted genes. We found that these three miRNAs are involved in many crucial signaling pathways, including adherens junction, autophagy, and TGF-beta, FoxO, and ErbB signaling pathway. Moreover, accumulating evidences showed that autophagy was correlated with the progression of breast cancer, including proliferation, invasion and metastasis (34). TGF-β was involved in various biological functions including cell differentiation, metastasis, proliferation, angiogenesis, and cellular microenvironment (35). Inhibition of TGF-β pathway is a potential new therapeutic strategy for breast cancer (36). In addition, Bullock et al. reported that the PI3K/AKT/FOXO signaling axis plays a crucial role in the hormone-independent growth of many breast cancers (37). However, further experiments are needed to verify these predictions.
In conclusion, we found that a three-miRNA signature could validly predict the prognosis of the patients with breast cancer. Nevertheless, further functional experiments are required to verify the biological functions of this signature in breast cancer progression.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.02.21). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Institutional ethical approval and informed consent were waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Li Y, Meeran SM, Patel SN, et al. Epigenetic reactivation of estrogen receptor-alpha (ERalpha) by genistein enhances hormonal therapy sensitivity in ERalpha-negative breast cancer. Mol Cancer 2013;12:9. [Crossref] [PubMed]
- Chen L, Kang C. miRNA interventions serve as 'magic bullets' in the reversal of glioblastoma hallmarks. Oncotarget 2015;6:38628-42. [PubMed]
- Hou LK, Ma YS, Han Y, et al. Association of microRNA-33a Molecular Signature with Non-Small Cell Lung Cancer Diagnosis and Prognosis after Chemotherapy. PLoS One 2017;12:e0170431. [Crossref] [PubMed]
- Tian XP, Wang CY, Jin XH, et al. Acidic Microenvironment Up-Regulates Exosomal miR-21 and miR-10b in Early-Stage Hepatocellular Carcinoma to Promote Cancer Cell Proliferation and Metastasis. Theranostics 2019;9:1965-79. [Crossref] [PubMed]
- Fu Y, Wang C, Zhang D, et al. miR-15b-5p ameliorated high glucose-induced podocyte injury through repressing apoptosis, oxidative stress, and inflammatory responses by targeting Sema3A. J Cell Physiol 2019;234:20869-78. [Crossref] [PubMed]
- Liu F, Sang M, Meng L, et al. miR92b promotes autophagy and suppresses viability and invasion in breast cancer by targeting EZH2. Int J Oncol 2018;53:1505-15. [PubMed]
- Pillman KA, Phillips CA, Roslan S, et al. miR-200/375 control epithelial plasticity-associated alternative splicing by repressing the RNA-binding protein Quaking. EMBO J 2018;37:e99016. [Crossref] [PubMed]
- Pesce S, Squillario M, Greppi M, et al. New miRNA Signature Heralds Human NK Cell Subsets at Different Maturation Steps: Involvement of miR-146a-5p in the Regulation of KIR Expression. Front Immunol 2018;9:2360. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Zampini M, Bisio V, Leszl A, et al. A three-miRNA-based expression signature at diagnosis can predict occurrence of relapse in children with t(8;21) RUNX1-RUNX1T1 acute myeloid leukaemia. Br J Haematol 2018;183:298-301. [Crossref] [PubMed]
- Chu CA, Lee CT, Lee JC, et al. MiR-338-5p promotes metastasis of colorectal cancer by inhibition of phosphatidylinositol 3-kinase, catalytic subunit type 3-mediated autophagy pathway. EBioMedicine 2019;43:270-81. [Crossref] [PubMed]
- Jin D, Guo J, Wu Y, et al. UBE2C, Directly Targeted by miR-548e-5p, Increases the Cellular Growth and Invasive Abilities of Cancer Cells Interacting with the EMT Marker Protein Zinc Finger E-box Binding Homeobox 1/2 in NSCLC. Theranostics 2019;9:2036-55. [Crossref] [PubMed]
- Hoareau-Aveilla C, Quelen C, Congras A, et al. miR-497 suppresses cycle progression through an axis involving CDK6 in ALK-positive cells. Haematologica 2019;104:347-59. [Crossref] [PubMed]
- Huang L, Tang X, Shi X, et al. miR-532-5p promotes breast cancer proliferation and migration by targeting RERG. Exp Ther Med 2020;19:400-8. [PubMed]
- Sun Q, Shen X, Wang P, et al. Targeting cyclophilin-D by miR-1281 protects human macrophages from Mycobacterium tuberculosis-induced programmed necrosis and apoptosis. Aging (Albany NY) 2019;11:12661-73. [Crossref] [PubMed]
- Mammadzada P, Bayle J, Gudmundsson J, et al. Identification of Diagnostic and Prognostic microRNAs for Recurrent Vitreous Hemorrhage in Patients with Proliferative Diabetic Retinopathy. J Clin Med 2019;8:2217. [Crossref] [PubMed]
- Seo HA, Moeng S, Sim S, et al. MicroRNA-Based Combinatorial Cancer Therapy: Effects of MicroRNAs on the Efficacy of Anti-Cancer Therapies. Cells 2019;9:29. [Crossref] [PubMed]
- Zhou W, Fong MY, Min Y, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014;25:501-15. [Crossref] [PubMed]
- Jin X, Yu Y, Zou Q, et al. MicroRNA-105 promotes epithelial-mesenchymal transition of nonsmall lung cancer cells through upregulating Mcl-1. J Cell Biochem 2019;120:5880-8. [Crossref] [PubMed]
- Wu D, Chen B, Cui F, et al. Hypoxia-induced microRNA-301b regulates apoptosis by targeting Bim in lung cancer. Cell Prolif 2016;49:476-83. [Crossref] [PubMed]
- Fan H, Jin X, Liao C, et al. MicroRNA-301b-3p accelerates the growth of gastric cancer cells by targeting zinc finger and BTB domain containing 4. Pathol Res Pract 2019;215:152667. [Crossref] [PubMed]
- Xia X, Zhang K, Luo G, et al. Downregulation of miR-301a-3p sensitizes pancreatic cancer cells to gemcitabine treatment via PTEN. Am J Transl Res 2017;9:1886-95. [PubMed]
- Zhou SL, Yin D, Hu ZQ, et al. A Positive Feedback Loop Between Cancer Stem-Like Cells and Tumor-Associated Neutrophils Controls Hepatocellular Carcinoma Progression. Hepatology 2019;70:1214-30. [Crossref] [PubMed]
- Stevic I, Muller V, Weber K, et al. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Med 2018;16:179. [Crossref] [PubMed]
- Guo YJ, Liu JX, Guan YW. Hypoxia induced upregulation of miR-301a/b contributes to increased cell autophagy and viability of prostate cancer cells by targeting NDRG2. Eur Rev Med Pharmacol Sci 2016;20:101-8. [PubMed]
- Wang W, Liu M, Guan Y, et al. Hypoxia-Responsive Mir-301a and Mir-301b Promote Radioresistance of Prostate Cancer Cells via Downregulating NDRG2. Med Sci Monit 2016;22:2126-32. [Crossref] [PubMed]
- Song H, Li D, Wu T, et al. MicroRNA-301b promotes cell proliferation and apoptosis resistance in triple-negative breast cancer by targeting CYLD. BMB Rep 2018;51:602-7. [Crossref] [PubMed]
- Zhang L, Sullivan PS, Goodman JC, et al. MicroRNA-1258 suppresses breast cancer brain metastasis by targeting heparanase. Cancer Res 2011;71:645-54. [Crossref] [PubMed]
- Zhang H, Jiang S, Guo L, et al. MicroRNA-1258, regulated by c-Myb, inhibits growth and epithelial-to-mesenchymal transition phenotype via targeting SP1 in oral squamous cell carcinoma. J Cell Mol Med 2019;23:2813-21. [Crossref] [PubMed]
- Zhang Z, Li J, Huang Y, et al. Upregulated miR-1258 regulates cell cycle and inhibits cell proliferation by directly targeting E2F8 in CRC. Cell Prolif 2018;51:e12505. [Crossref] [PubMed]
- Jiang W, Wei K, Pan C, et al. MicroRNA-1258 suppresses tumour progression via GRB2/Ras/Erk pathway in non-small-cell lung cancer. Cell Prolif 2018;51:e12502. [Crossref] [PubMed]
- Liu W, Zhou Z, Zhang Q, et al. Overexpression of miR-1258 inhibits cell proliferation by targeting AKT3 in osteosarcoma. Biochem Biophys Res Commun 2019;510:479-86. [Crossref] [PubMed]
- Li M, Liu J, Li S, et al. Autophagy-related 7 modulates tumor progression in triple-negative breast cancer. Lab Invest 2019;99:1266-74. [Crossref] [PubMed]
- Syed V. TGF-beta Signaling in Cancer. J Cell Biochem 2016;117:1279-87. [Crossref] [PubMed]
- Khoshakhlagh M, Soleimani A, Binabaj MM, et al. Therapeutic potential of pharmacological TGF-beta signaling pathway inhibitors in the pathogenesis of breast cancer. Biochem Pharmacol 2019;164:17-22. [Crossref] [PubMed]
- Bullock M. FOXO factors and breast cancer: outfoxing endocrine resistance. Endocr Relat Cancer 2016;23:R113-30. [Crossref] [PubMed]