
Candidate genes for predicting the survival of patients with gastric cancer: a study based on The Cancer Genome Atlas (TCGA) database
Introduction
Gastric cancer (GC) is the second most frequent cause of cancer-related mortality in the world. Despite the developments in endoscopic technology, the great progression made in early cancer screening, and the achievements made in relation to Helicobacter pylori eradication, the 5-year survival rate for GC remains very low worldwide (1).
Dynamic changes in the genome play an essential role in the progress of carcinogenesis (2). The Cancer Genome Atlas (TCGA) provides a comprehensive overview of gene expression, RNA-seq, DNA copy-number, somatic mutations, and DNA methylation profiles in tumors, as well as providing the matched clinical information of patients with cancer (3). This publicly available cancer genomics data set allows for improved diagnostic methods, treatment criteria and, ultimately, cancer prevention (4).
Many studies have proved that, in GC patients, the TNM stage is not the only factor impacting survival (5); gene expression also bears a strong association. Previous studies have revealed that the overexpression of tumor protein 53 (p53) and Mucin 1 (MUC1), and the decrease of expressions of phosphatase and tension homolog gene (PTEN), E-cadherin gene, and SMAD family member 4 (SMAD4), were found to be associated with poor prognosis of GC patients (6). Recent studies have also found that people with high expression of LncRNA AL139147 show a tendency towards poor prognosis (7). Competing endogenous RNAs (ceRNA) analysis has also shown that the complex mechanisms of the ceRNA network are essential in the progression of GC (8). Various genes that could be considered as candidate prognostic biomarkers and therapeutic targets are yet to be revealed and comprehensively understood.
In this study, we obtained the gene expression profiles of 375 gastric tumors and 32 adjacent non-tumor samples from TCGA database. A Gene expression matrix was obtained, and R package “edgeR” was used to examine differentially expressed genes (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html) (9). The gene expression profiles were combined with clinical survival information. Integrated bioinformatic analyses were performed using “R”, including differential gene expression analysis, The Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis (10,11), as well as survival analysis. To identify which gene could predict the outcomes of GC patients, Kaplan-Meier survival analysis was performed.
Methods
Gene expression profile and patient clinical data
We downloaded the gene expression profiles and clinical data of GC patients from TCGA database (https://cancergenome.nih.gov/) in November 2018 and analyzed the statistics between December 2018 and May 2019. The exclusion criteria were as follows: (I) without clinical information or prognostic statistics like survival time; (II) without matched adjacent non-tumor tissues; and (III) not stomach adenocarcinoma. Ultimately, 407 samples including 375 GC tumor tissues and 32 adjacent non-tumor samples, were collected for integrated bioinformatics analysis. There was no need for ethical approval as all data in this study were downloaded from public databases (TCGA), and the data processing met the TCGA publication guidelines (https://cancergenome.nih.gov/publications/guidelines).
Differential gene expression in GC
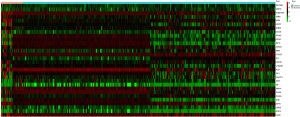
The gene expression profiles of tumor tissue and adjacent non-tumor tissue samples from GC patients were analyzed in R using the “edgeR” package and were normalized by log2 transformation. We used fold change (FC) to characterize the expression differences. Each gene has its associated P values. The “edgeR” package was used to determine the differentially expressed genes with a cutoff of P<0.05 and |logFC |>2 to define the differential expression of genes in GC patients. The unbiased t-test provided by the “Limma” package in “R” was used to evaluate the significant P value of differences in gene expression (12). All the genes were tested by t-test to determine their corresponding P value. A heat map of the top 30 differentially expressed genes were drawn by the “pheatmap” package in “R” (13). The heat map was divided into two categories, the tumor tissue group and the adjacent nontumor tissue group. Red represents the up-regulation of gene expression, and the green represents the down-regulation of gene expression.
Functional enrichment analysis
To better understand the biological functions of the dysregulated genes, GO biological enrichment and KEGG pathways analysis were performed through the “ggplot2” and “clusterProfiler” package in “R” (14). DAVID database was used to carry out functional enrichment analysis (https://david-d.ncifcrf.gov/) (15). GO analysis results included three parts, biological process (BP), cellular component (CC), and molecular function (MF). P<0.05 was considered significant.
Survival analysis to search for the candidate genes
GC samples were divided into two groups according to gene expression: the high expression group and the low expression group. Kaplan-Meier survival analysis was conducted using the “survival” package in “R” to explore the associations between the expression of a specific gene and prognosis of GC patients. We analyzed the top 30 differentially expressed genes from 1,313 genes with expression differences, as well as all the genes enriched in the top 29 KEGG pathways, ranked by P value. The log-rank test was used to determine significant differences in survival curves (16), and P value <0.05 was considered as statistically significant.
Results
Identification of mRNAs in GC
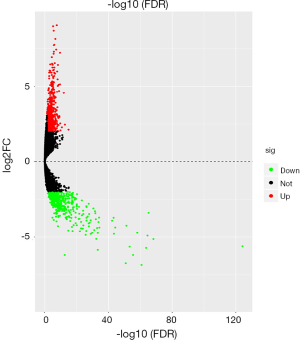
A total of 407 samples, including 375 GC tumor tissue samples and 32 adjacent nontumor tissue samples, were collected for this study. There were 1,313 differentially expressed genes in total, including 781 up-regulated and 532 down-regulated genes identified in GC and matched normal tissues. The cut-off criteria of differentially expressed genes was P<0.05 and |logFC| >2. The volcano plot of the differentially expressed genes is presented in Figure 1. The red dots represent the up-regulated genes, while the green dots represent the down-regulated genes. The heat map of the top 30 differentially expressed genes ranked according to the fold change was conducted in R with the package “pheatmap” (https://cran.Rproject.org/web/packages/pheatm/apindex.html) and is shown in Figure 2.
Functional analysis
To better understand the genes’ function, Gene Oncology (GO) and Kyoto Encyclopedia of Genes and Genomics (KEGG) analysis were performed in “R”. The up-regulated and down-regulated genes were separately analyzed by KEGG analysis. The top 29 terms with the lowest P value were selected. The results (Figure 3, Table 1) showed that the down-regulated genes were significantly enriched in pathways such as the cGMP-PKG, estrogen, and cAMP signaling pathways, as well as the PPAR signaling pathway. The down-regulated genes also interacted with gastric acid secretion, protein digestion and absorption, and insulin secretion. The up-regulated genes were primarily enriched in cytokine-cytokine receptor interaction. The top six GO terms (Figure 4, Table 2) were “digestion”, “peptide cross-linking”, “keratinocyte differentiation”, “erythrocyte differentiation”, “proteolysis”, and “detection of chemical stimulus involved in sensory perception of the bitter taste”. GO analysis results included BP, CC, and MF, and P<0.05 was considered as statistically significant.
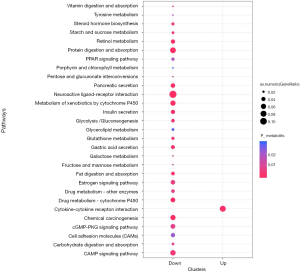
Table 1
| Cluster | Description | P value | Count | Gene |
|---|---|---|---|---|
| Down | Protein digestion and absorption | 3.11E−09 | 16 | MEP1B/PGA3/CELA3A/COL4A6/CELA2A/MME/ATP1A2/CPB1/PGA4/SLC15A1/MEP1A/PGA5/XPNPEP2/SLC6A19/SLC7A9/SLC8A2 |
| Down | Metabolism of xenobiotics by cytochrome P450 | 1.58E−07 | 13 | CYP3A4/AKR1C1/ALDH3A1/GSTA2/ADH7/UGT1A7/GSTA1/GSTM5/ADH4/UGT2B17/UGT1A5/ADH1B/CBR1 |
| Down | Chemical carcinogenesis | 3.97E−07 | 13 | CYP3A4/ALDH3A1/GSTA2/ADH7/UGT1A7/GSTA1/GSTM5/ADH4/UGT2B17/UGT1A5/AKR1C2/ADH1B/CBR1 |
| Down | Drug metabolism-cytochrome P450 | 4.62E−06 | 11 | CYP3A4/ALDH3A1/GSTA2/ADH7/UGT1A7/GSTA1/GSTM5/ADH4/UGT2B17/UGT1A5/ADH1B |
| Down | Neuroactive ligand-receptor interaction | 8.53E−06 | 25 | GRIK5/GIP/CORT/LPAR3/GRIA2/GABRA5/TACR2/HRH3/GHRL/PENK/GRIA4/GALR1/VIP/NPY2R/AVPR1B/ADCYAP1/SST/GRPR/GHR/CCKAR/LEP/GCG/P2RY14/GRP/PTH2R |
| Down | Retinol metabolism | 9.82E−05 | 9 | CYP3A4/ADH7/UGT1A7/RDH12/BCO1/ADH4/UGT2B17/UGT1A5/ADH1B |
| Down | Fat digestion and absorption | 0.000127444 | 7 | MTTP/APOB/LIPF/MOGAT2/FABP1/APOA1/ABCG5 |
| Down | Pancreatic secretion | 0.000412655 | 10 | CELA3A/CELA2A/ATP1A2/CPB1/ATP2B3/CLCA1/CLCA4/CCKAR/SLC26A3/KCNMA1 |
| Down | Insulin secretion | 0.000665041 | 9 | GIP/CREB3L3/ATP1A2/CACNA1S/ADCYAP1/CCKAR/GCG/KCNMB1/KCNMA1 |
| Down | Gastric acid secretion | 0.001163971 | 8 | KCNE2/ATP4B/ATP4A/ATP1A2/SST/SLC9A4/MYLK/GAST |
| Down | cAMP signaling pathway | 0.002601643 | 14 | GIP/CREB3L3/GRIA2/ATP1A2/PLN/GHRL/ATP2B3/GRIA4/VIP/CACNA1S/MYL9/ADCYAP1/SST/GCG |
| Down | Glycolysis/gluconeogenesis | 0.002889032 | 7 | ALDOB/ALDH3A1/ADH7/PCK1/G6PC/ADH4/ADH1B |
| Down | Starch and sucrose metabolism | 0.003138338 | 5 | MGAM/TREH/ENPP3/MGAM2/G6PC |
| Down | Vitamin digestion and absorption | 0.004184031 | 4 | RBP2/APOB/GIF/APOA1 |
| Down | Glutathione metabolism | 0.004686566 | 6 | NAT8/GSTA2/GSTA1/GPX3/GSTM5/ANPEP |
| Down | Estrogen signaling pathway | 0.005488775 | 10 | KRT37/ESR1/TFF1/CREB3L3/GNAO1/KRT15/HSPA2/PGR/KRT20/KRT13 |
| Down | Steroid hormone biosynthesis | 0.006585195 | 6 | CYP3A4/AKR1C1/UGT1A7/UGT2B17/UGT1A5/AKR1C2 |
| Down | Drug metabolism-other enzymes | 0.00666097 | 7 | CYP3A4/GSTA2/UGT1A7/GSTA1/GSTM5/UGT2B17/UGT1A5 |
| Down | cGMP-PKG signaling pathway | 0.007128674 | 11 | CREB3L3/ATP1A2/PLN/ATP2B3/CACNA1S/MYL9/SLC8A2/MYLK/KCNMB1/RGS2/KCNMA1 |
| Down | Carbohydrate digestion and absorption | 0.007537726 | 5 | MGAM/MGAM2/SLC2A5/ATP1A2/G6PC |
| Down | Galactose metabolism | 0.01062647 | 4 | MGAM/MGAM2/G6PC/AKR1B10 |
| Down | Fructose and mannose metabolism | 0.013229258 | 4 | ALDOB/PFKFB1/KHK/AKR1B10 |
| Down | Pentose and glucuronate interconversions | 0.014669893 | 4 | UGT1A7/UGT2B17/UGT1A5/AKR1B10 |
| Down | PPAR signaling pathway | 0.017632254 | 6 | RXRG/APOC3/PLIN4/PCK1/FABP1/APOA1 |
| Down | Tyrosine metabolism | 0.017839499 | 4 | ALDH3A1/ADH7/ADH4/ADH1B |
| Down | Cell adhesion molecules (CAMs) | 0.021986616 | 9 | CD276/CLDN14/NRXN1/CLDN23/MPZ/CLDN17/NEGR1/CNTN1/NRXN3 |
| Down | Glycerolipid metabolism | 0.028159835 | 5 | LIPF/MOGAT2/DGKB/AKR1B10/PNLIPRP3 |
| Down | Porphyrin and chlorophyll metabolism | 0.029764972 | 4 | UGT1A7/COX10/UGT2B17/UGT1A5 |
| Up | Cytokine-cytokine receptor interaction | 0.000755068 | 22 | CXCL8/IL13RA2/IFNL2/IL15RA/CXCL10/INHBA/CXCL9/PF4V1/CXCL6/GDF1/CXCL11/TNFSF11/TNFRSF11B/PPBP/THPO/CXCL5/BMP8A/IL17F/CCL26/GDF15/CCL3/CSH2 |
Count: the number of enriched genes in each term.
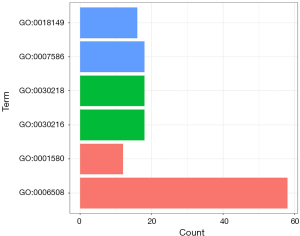
Table 2
| Category | Term | Name | Count | Percent | P value | FDR |
|---|---|---|---|---|---|---|
| MF | GO:0007586 | Digestion | 18 | 1.417322835 | 0.000000231 | 0.00042 |
| MF | GO:0018149 | Peptide cross-linking | 16 | 1.25984252 | 0.00000026 | 0.000472 |
| CC | GO:0030216 | Keratinocyte differentiation | 18 | 1.417322835 | 0.00000399 | 0.007254384 |
| CC | GO:0030218 | Erythrocyte differentiation | 18 | 1.417322835 | 0.00000399 | 0.007254384 |
| BP | GO:0006508 | Proteolysis | 58 | 4.566929134 | 0.0000111 | 0.020147299 |
| BP | GO:0001580 | Detection of chemical stimulus involved in sensory perception of bitter taste | 12 | 0.94488189 | 0.0000259 | 0.047106424 |
Count: the number of enriched genes in each term. BP, biological process; CC, cellular component; MF, molecular function.
Survival analysis
To ascertain which candidate genes may influence survival outcomes in GC patients, survival analysis was performed using the Kaplan-Meier method with a log-rank statistical test. The patients with GC were categorized into a high-expression group and a low-expression group according to its median gene expression level. We downloaded the survival information of the samples from the TCGA database and obtained the matched survival status of each sample. We analyzed the top 30 differentially expressed genes using the “survival” package in “R”, ranked according to the fold change, we also analyzed all the genes enriched in the top 29 KEGG pathway, ranked by P value.
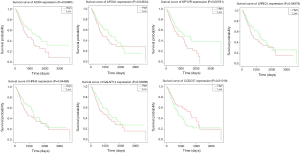
Seven genes were found to be associated with survival: ADH4, HRH3, NPY2R, APOA1, GALNT14, LRRIQ1, and CCDC57. A significance level of P<0.05 was set as the cut-off criteria, and the results are shown in Figure 5. GALNT14, LRRIQ1, and CCDC57 were selected as candidate genes from the top 30 differentially expressed genes, and ADH4, HRH3, NPY2R, and APOA1 were associated with the top 29 KEGG pathways: “Metabolism of xenobiotics by cytochrome P450”, “Chemical carcinogenesis”, “Drug metabolism-cytochrome P450” “Retinol metabolism”, “Glycolysis/Gluconeogenesis” and “Tyrosine metabolism”, HRH3 and NPY2R were found to be linked with the neuroactive ligand-receptor interaction pathway. APOA1 interacted with the fat digestion and absorption, vitamin digestion and absorption, and PPAR signaling KEGG pathways. GC patients who had high expression of LRRIQ1, GALNT14, APOA1, NPY2R, HRH3, and ADH4 had a better prognosis than GC patients with low expressions of these genes, while the GC patients with low expression of CCDC57 often had poor survival outcomes.
Discussion
GC is one of the most malignant cancers worldwide, although great progress has been made in endoscopic surveillance for early GC, and many new molecular targeted drugs have been invented and clinically applied, such as the human epidermal growth factor receptor 2 (HER-2)-targeted drug trastuzumab (17). In spite of this, the 5-year survival rate remains low (29.6%) for GC patients around the world (1). Many genes are overexpressed in GC, and some of these genes could be potential prognosis predictors and/or therapeutic targets. It has been proved that the accumulation of mutations in crucial genes may cause cancer by altering normal programs of differentiation and cell proliferation and death (18). Genetic changes often lead to the alteration of biological processes. TCGA project wants to identify dysregulated pathways and candidate driver genes in GC (19).
In this study, we conducted some bioinformatic analyses to determine the candidate genes which can predict survival in GC patients. At first, we found a total of 1,313 differentially expressed genes, including 781 up-regulated and 532 down-regulated genes. Among the top 30 differentially expressed genes and all the differentially expressed genes enriched in the top 29 KEGG pathways, 7 genes (ADH4, HRH3, NPY2R, APOA1, GALNT14, LRRIQ1, and CCDC57) were selected as the candidate genes. GC patients with low expression of CCDC57 often had poor survival outcomes. GC patients with low expression of any one of the other six genes (ADH4, HRH3, NPY2R, APOA1, GALNT14, and LRRIQ1) often had a good survival outcome.
acetylgalactosaminyltransferase 14 (GALNT14) belongs to the polypeptide N-acetylgalactosaminyltransferase family. Previous studies found that the loss function of GALNTs can result in altered glycoproteins and can cause tumor aggressiveness in various kinds of cancer (20). The genotype of polypeptide GALNT14 has also be put forward as a potential prognostic predictor for patients undergoing chemotherapy for hepatocellular carcinoma (21). The human alcohol dehydrogenase 4 gene (ADH4) is a member of the human alcohol dehydrogenase (ADH) family, which plays a role in the process of ethanol metabolism (22). Neuropeptide Y (NPY) is an appetite hormone that has been reported to be a candidate gene associated with the development of obesity and control of food intake (23-27). Apolipoprotein AI (APOA1) belongs to the apolipoprotein family (28). By using gene expression array analysis, it has been found that APOA1 mRNA expression in ovarian serous is a marker of longer survival (29). A retrospective study involving 1,201 GC patients who received surgery showed that patients with high ApoB1/ApoA1 (≥1) had shorter overall survival (30). The histamine receptor H3 (HRH3) has been identified as an important molecule in inflammation and carcinogenesis. Recent studies have found that HRH4 is involved in inflammation-related colorectal carcinogenesis (31). Coiled-coil-domain-containing 57 (CCDC57) has been found to be slightly higher in uterine leiomyomata (32). Previous studies have found that the methylation of CCDC57 is related to age, tumor location, and Helicobacter infection in early gastric carcinogenesis (33). Our study shows that these genes are associated with the outcomes of GC patients, but their molecular mechanisms are still poorly understood.
In summary, we selected seven candidate genes that could be considered as candidate prognostic biomarkers in GC patients. These seven genes may become future therapeutic targets in GC. However, our study needs another validation cohort to verify our results, and further investigation and molecular experiments are required to explore the roles of these genes in GC better.
Acknowledgments
Funding: This work is supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.02.82). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57-70. [Crossref] [PubMed]
- Kim HS, Minna JD, White MA. GWAS meets TCGA to illuminate mechanisms of cancer predisposition. Cell 2013;152:387-9. [Crossref] [PubMed]
- Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68-77. [Crossref] [PubMed]
- de jesus VHF, da costa WL, Felismino TC, et al. Survival outcomes of patients with pathological stage I gastric cancer using the competing risks survival method. J Gasreointest Oncol 2019;10:1110-9.
- Lee HS, Lee HK, Kim HS, et al. Tumour suppressor gene expression correlates with gastric cancer prognosis. J Pathol 2003;200:39-46. [Crossref] [PubMed]
- Li F, Huang C, Li Q, et al. Construction and analysis of lncRNA-associated ceRNA network identified potential prognostic biomarker in gastric cancer. Med Sci Monit 2018;24:37-49. [Crossref] [PubMed]
- Yang XZ, Cheng TT, He QJ, et al. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol Cancer 2018;17:126. [Crossref] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Smyth GK. limma: Linear Models for Microarray Data.
- Wang L, Cao C, Ma Q, et al. RNA-seq analyses of multiple meristems of soybean: novel and alternative transcripts, evolutionary and functional implications. BMC Plant Biol 2014;14:169. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 2012;16:284-7. [Crossref] [PubMed]
- Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Bewick V, Cheek L, Ball J. Statistics review 12: survival analysis. Crit Care 2004;8:389-94. [Crossref] [PubMed]
- Boku N. HER2-positive gastric cancer. Gastric Cancer 2014;17:1-12. [Crossref] [PubMed]
- Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949-54. [Crossref] [PubMed]
- Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- De Mariano M, Gallesio R, Chierici M, et al. Identification of GALNT14 as a novel neuroblastoma predisposition gene. Oncotarget 2015;6:26335-46. [Crossref] [PubMed]
- Liang KH, Lin CC, Yeh CT. GALNT14 SNP as a potential predictor of response to combination chemotherapy using 5-FU, mitoxantrone and cisplatin in advanced HCC. Pharmacogenomics 2011;12:1061-73. [Crossref] [PubMed]
- Osier M, Pakstis AJ, Kidd JR, et al. Linkage disequilibrium at the ADH2 and ADH3 loci and risk of alcoholism. Am J Hum Genet 1999;64:1147-57. [Crossref] [PubMed]
- Campbell CD, Lyon HN, Nemesh J, et al. Association studies of BMI and type 2 diabetes in the neuropeptide Y pathway: a possible role for NPY2R as a candidate gene for type 2 diabetes in men. Diabetes 2007;56:1460-7. [Crossref] [PubMed]
- Torekov SS, Larsen LH, Andersen G, et al. Variants in the 5' region of the neuropeptide Y receptor Y2 gene (NPY2R) are associated with obesity in 5,971 white subjects. Diabetologia 2006;49:2653-8. [Crossref] [PubMed]
- Siddiq A, Gueorguiev M, Samson C, et al. Single nucleotide polymorphisms in the neuropeptide Y2 receptor (NPY2R) gene and association with severe obesity in French white subjects. Diabetologia 2007;50:574-84. [Crossref] [PubMed]
- Wang HJ, Wermter AK, Nguyen TT, et al. No association of sequence variants in the neuropeptide Y2 receptor (NPY2R) gene with early onset obesity in Germans. Horm Metab Res 2007;39:840-4. [Crossref] [PubMed]
- Hunt SC, Hasstedt SJ, Xin Y, et al. Polymorphisms in the NPY2R gene show significant associations with BMI that are additive to FTO, MC4R, and NPFFR2 gene effects. Obesity 2011;19:2241-7. [Crossref] [PubMed]
- Hamon SC, Kardia SL, Boerwinkle E, et al. Evidence for consistent intragenic and intergenic interactions between SNP effects in the APOA1/C3/A4/A5 gene cluster. Hum Hered 2006;61:87-96. [Crossref] [PubMed]
- Tuft Stavnes H, Nymoen DA, Hetland Falkenthal TE, et al. APOA1 mRNA expression in ovarian serous carcinoma effusions is a marker of longer survival. Am J Clin Pathol 2014;142:51-7. [Crossref] [PubMed]
- Ma MZ, Yuan SQ, Chen YM, et al. Preoperative apolipoprotein B/apolipoprotein A1 ratio: a novel prognostic factor for gastric cancer. Onco Targets Ther 2018;11:2169-76. [Crossref] [PubMed]
- Tanaka T, Kochi T, Shirakami Y, et al. Cimetidine and Clobenpropit Attenuate Inflammation-Associated Colorectal Carcinogenesis in Male ICR Mice. Cancers (Basel) 2016;8:25. [Crossref] [PubMed]
- Eggert SL, Huyck KL, Somasundaram P, et al. Genome-wide linkage and association analyses implicate FASN in predisposition to Uterine Leiomyomata. Am J Hum Genet 2012;91:621-8. [Crossref] [PubMed]
- Chong Y, Mia-Jan K, Ryu H, et al. DNA methylation status of a distinctively different subset of genes is associated with each histologic Lauren classification subtype in early gastric carcinogenesis. Oncol Rep 2014;31:2535-44. [Crossref] [PubMed]


