Exploration of the genomic landscape of a long-term surviving stage III colorectal cancer patient identifies recurrent and rare mutations: a case report
Introduction
Next-generation sequencing technologies have significantly transformed cancer genomics research through provision of reliable information about individual tumors, paving ways for precision therapy (1). WGS enhances the sequencing of the entire cancer patient genome and can be employed to detect all germline and somatic mutations, including copy number alternations, gene fusions and chromosomal rearrangements (2). Colorectal cancer (CRC) is one of the leading causes of cancer-associated mortality worldwide with about 1.8 million new cases and 860,000 deaths reported in 2018 (3). CRC is less common in young individuals and often times, young patients with CRC present with late-stage disease (stage III or IV) (4). Stage III patients constitute a considerable proportion of most CRCs, and account for about one-third of all reported cases (5). More than two-thirds of all colorectal patients undergo curative surgery, with a considerable percentage of stage III patients experiencing tumor relapse manifesting as metastasis to distant organs or metachronous colorectal lesions within 5 years of follow up (6). Despite increasing scientific evidences that CRC is a heterogeneous disease, and genetic characteristics of the tumors influence patient prognosis and response to targeted therapies (7), the exploration of individual mutation profiles of same stage, advanced CRC patients has not received significant attention. We here explored the genomic landscape of a young long-term surviving stage III CRC patient and compared our data with those of conventional stage III patients reported in The Cancer Genome Atlas Network (TCGA) database. Mutations in APC, TP53, KRAS, SMAD4, FBXW7 and PIK3CA observed in TCGA patients were not recorded in our study. However, mutations in MUC4, MUC16, ARID1B, BAZ1A, BRCA2, CTNND1 and NCOA2 rarely reported in TCGA patients were predominant in our patient. Additionally, we observed loss of heterozygosity (LOH) in POLE, RET, BMPR1A, NCOA4 and 30 other genes in contrast to deletion and amplification events recorded in TCGA patients. Put together, we offered substantial insights into the genomic features of the patient and provided a valuable resource for further study into the mutations that characterize advanced CRC which may be useful to design clinical therapy for personalized medicine. We present the following case in accordance with the CARE Guideline.
Case presentation
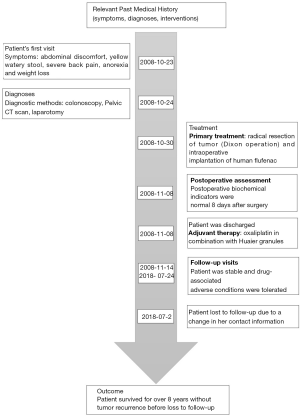
A 40-year-old female patient presented CRC related symptoms and was admitted to the First Affiliated Anhui Medical University on October 23, 2008. She is a primary school teacher; has harmonious social relations and is married with two kids. The onset of symptoms was 4-month prior to her admission and this included abdominal discomfort, yellow watery stool and severe back pain. She had previously taken some self-prescribed intestinal drugs which made the symptoms subside. Much later, she developed diarrhea, and her tenesmus got worse. Although her body temperature and urine were normal, she experienced abdominal distension and pain. She also became anorexic and began to lose weight. She however denied any known family history of cancer. Diagnosis through colonoscopy revealed she had rectal invasive moderately differentiated adenocarcinoma. Her tumor size was 3 cm × 3 cm, and the carcinoma had invaded the serosa layer, covering 3/4 of the intestine tube. Upon examination of the mesenteric, two of her four lymph nodes showed metastases, although no obviously enlarged lymph nodes were found in the mesenteric roots. The patient was diagnosed with a stage III (T3N1M0, IIB) CRC. Examination of the rectum left lateral position showed there was a smooth mass 4 cm from the verge of the anus. The boundary was clear and the mass had a size of 2 cm × 2 cm. Pelvic CT scan revealed that the bowel was markedly narrowed about 10 cm from the anus and the site was seen as an irregular tissue shadow, with a length of 4 cm. From the colonoscopy, infiltrating lesion with four walls was observed at 12 cm, with an ulcerated surface. After laparotomy, the mass was found on the anterior wall of the rectum and the peritoneal reflex, 8 cm from the anal verge. Radical resection (Dixon operation) of the tumor was performed under anesthesia with intraoperative implantation of 600 mg of human flufenac into her abdominal cavity. Oxaliplatin in combination with Huaier granules, a traditional Chinese medicine, was given as adjuvant therapy. Huaier granule was applied in this case due to its affordability and demonstrated anti-tumor effects in various cancer including CRC. The clinical validity of Huaier granules combined with chemotherapy as adjuvant therapy after curative surgery in CRC patients has been demonstrated in several studies (8-10). Follow-up examination showed that the biochemical index was normal at the 8th day after surgery; the Neutrophils were slightly higher, while the lymphocytes were a little lower. The patient received relevant treatments under the guidance of a Doctor in compliance with the treatment plan and there were no adverse reactions to the treatment regimen. The patient had an overall survival of over 8-year and was lost to follow-up in July 2018 due to a change in her contact information (see Figure 1 for medical history timeline). To obtain the mutation profile of the patient, genomic DNA was isolated from the Formalin-Fixed, Paraffin-Embedded (FFPE) tumor and matched adjacent normal tissues using genomic DNA isolation kit (QIAGEN, Hilden, Germany). Extracted DNA was quantified using a NanoDrop ND-1000 spectrophotometer and the integrity was assessed with agarose gel electrophoresis. WGS of the prepared libraries was performed using the Illumina X10 (Illumina Inc., San Diego, CA, USA) with 150-bp paired-end reads. Germline variants were called with GATK HaplotyperCaller joint v3.8 (11). Variants which passed VQSR module, coverage ≥3 were retained while the variants showing genotype “./.” in samples were filtered. Three callers: Lancet, Mutect v1.1.4, and SomaticIndelDetector 2.3-9 (12) were employed for somatic variants calling. Variants from the three callers were merged and all variants that flag as “PASS” were kept. Further annotations for both germline and somatic variants were added to each mutation using ANNOVAR (13) and VEP (14), respectively. Several publicly available databases such as 1,000 Genome Project, the Exome Aggregation Consortium, the Genome Aggregation Database and the compiled scores prediction system dbnsfp33a were also employed. Somatic structural variations were identified using FACETS and DELLY (15), thereafter, duplication, deletion and hemizygous segments were kept and annotated with ENCODE gene symbol in R v3.5.1 (16). Functional effects of the germline mutations were assessed using Mutation Assessor, Mutation Taster, M-CAP, Polyphen2, PROVEAN and SIFT (17), and that of somatic mutations with VEP. Germline mutations predicted to be deleterious by at least two algorithms were considered potential driver mutations while genes predicted by VEP as either possibly or probably damaging were considered as potential driver genes. Germline mutation analysis revealed a total of 4,532,691 SNPs/Indels. Among these, 19,102 SNPs/Indels were detected in the exonic regions, of which 11,090 were predicted to be protein altering, including 8,413 non-synonymous SNVs, 951 frameshift deletions, 310 frameshift insertions, 542 non-frameshift deletions, 163 non-frameshift insertions, 224 stop-gain, and 9 stop-loss. We filtered the variants using a set of publicly available datasets to exclude >1% MAF variants reported in the 1,000 Genomes Project, the Exome Aggregation Consortium and the Genome Aggregation Database and got 194 germline variants including 20 previously reported in both the CGC and CPG databases (http://fp.amegroups.cn/cms/b0f2041ca135d69192fac2035a0f2398/tcr.2020.03.55-1.pdf). A total of 111 somatic variants were observed in the exonic regions including 8 frameshift deletion, 5 frameshift insertion, 6 in-frame deletion, 7 in-frame insertion, 72 missense, 2 nonsense and 11 splice site. Then, we filtered the variants with publicly available datasets to select the most informative somatic variants. Totally, we got 7 variants; ARID1B, BAZ1A, BRCA2, CTNND1, MUC16, MUC4, NCOA2 with perceived potential impact on CRC development. Functional annotations with VEP predicted MUC16 to be possibly damaging, while other genes were either benign or tolerated (Table 1). We got 1,996 structural events, including 1,367 translocations, 143 inversions, 306 tandem duplications and 180 deletions with DELLY; however, only in-frame events and fusions with distance over 50 Kb between two genes were kept. Ultimately, 53 significant structural events including 31 genes were reported (http://fp.amegroups.cn/cms/38b07ec6a6db1a6ef9d1ecafe7f379b4/tcr.2020.03.55-2.pdf). FACETS identified 107 large segments with copy number alternations, including 34 duplications, 12 loss of function and 69 hemizygous. Subsequently, we kept genes which were only alternated in coding sequences and got 34 genes with LOH and 1 with duplication. (http://fp.amegroups.cn/cms/02320f5ee071b3b895f6e8f14fcf2e75/tcr.2020.03.55-3.pdf). To investigate the differences between the mutations recorded in our patient and other stage III CRC cases, we queried TCGA database with the mutated genes. As we had an array of mutations, we focused our query on somatic mutations including copy number alterations. TCGA [colorectal adenocarcinoma (COAD)] comprises 8 cohort studies; DCFI Cell reports 2012, Genentech Nature 2012, TCGA Firehose Legacy, TCGA Nature 2012, TCGA PanCancer Atlas, MSKCC Genome Biology, 2014, MSKCC Cancer Cell, 2018, MSK Nature Medicine 2019 and contains mutation data of 3814 patients which cut across stages I to IV. For the database query, we excluded studies that did not provide information on patients’ age, tumor staging and survival outcomes. We also filtered out germline mutations as well as somatic mutations with unknown significance. However, we included somatic driver mutations annotated in OncoKB, cBioPortal and COSMIC. Of all the 4 databases queried, MUC16 predicted with potential driver mutations in our study, was modestly mutated in stage III patients reported in the DCFI Cell reports 2012 and TCGA Firehose Legacy studies. No profiles were returned for the MSKCC Cancer Cell 2018 project while higher mutation frequencies were recorded in TCGA PanCancer Atlas stage III patients. Furthermore, we straightly compared the exact different mutations in our patient with those of stage III CRC patients in TCGA database. Mutations in APC, TP53, KRAS, SMAD4, FBXW7 and PIK3CA predicted as drivers in TCGA stage III patients were not recorded in our study. However, mutations in MUC4, MUC16, ARID1B, BAZ1A, BRCA2, CTNND1 and NCOA2 rarely reported in TCGA patients were predominant in our patient. Additionally, we observed there were no clear-cut correlations between patient age and survival outcomes in TCGA stage III CRC patients as no consistent trend was observed for a given age group or survival time (http://fp.amegroups.cn/cms/1a4904747e13a11112c9f08435b73e79/tcr.2020.03.55-4.pdf; http://fp.amegroups.cn/cms/91f3dd625688c14169ffa9ed40a1d65e/tcr.2020.03.55-5.pdf). We queried the relevant data sets with the genes that showed copy number alternations in our study taking note to filter out germline mutations and copy number alterations of unknown significance. We observed LOH in POLE, RET, BMPR1A, NCOA4 and 30 other genes in contrast to deletion and amplification events recorded in TCGA stage III patients (http://fp.amegroups.cn/cms/b1237ee51a6b268f9c8278f795f21f71/tcr.2020.03.55-6.pdf; http://fp.amegroups.cn/cms/4845e2e4196a4c545b901f05c1e26832/tcr.2020.03.55-7.pdf). Besides the somatic mutations and CNAs, DELLY detected multiple structural variations in the BAGE2 gene. Although these variations were not captured in TCGA database, our literature search revealed that mutations in the gene is a rare occurrence in CRC patients.
Table 1
| Gene | Database | Chromosome | Variant classification |
SIFT | PolyPhen |
|---|---|---|---|---|---|
| ARID1B | CGC723 | chr6:157099313-157099315:CAC:- | In frame deletion | NSR | NSR |
| BAZ1A | CGC723 | chr14:35234140-35234140:C:T | Splice site | NSR | NSR |
| BRCA2 | CGC723_CPG114 | chr13:32912123-32912123:G:A | Missense Mutation | Tolerated (0.87) | Benign (0.011) |
| CTNND1 | CGC723 | chr11:57556564- 57556720:TTTTTGAATCTAGACTGGGCTGTTCTC TGTGTTAAACCAATCAGTTGCGACCTTCTCTTA ACAGGTGTGTATGGAAATATGTTTATTAAGAAG GAAAAATCTTACTTTTTAAGAAATATGTATTTTT; ATTCCTTTCATGTCATAGCAGAAAAAAATC:- | Splice site | NSR | NSR |
| MUC16 | CGC723 | chr19:9069747-9069747:G:A | Missense mutation | NSR | Probably damaging (0.981) |
| MUC4 | CGC723 | chr3:195509573-195509573:A:G | Missense mutation | Tolerated low confidence (0.28) | Benign (0.316) |
| NCOA2 | CGC723 | chr8:71069450-71069450:G:A | Missense mutation | Tolerated (0.16) | Benign (0.001) |
CRC, colorectal cancer; NSR, no score returned.
Discussion
Germline mutations analysis identified 20 SNPs/Indels reported in the CGC and CPG databases. The germline mutations ranged from recurrent cancer susceptibility genes with quantifiable risks to rare genes not conventionally associated with CRC (18,19). Recently, Gong et al. (20), profiled 618 multi-stage Chinese CRC patients including 226 stage III and reported pathogenic germline mutation in 1 of every 3 patients younger than 50, stressing the importance of genetic testing in all Chinese patients younger than 50. In our somatic SNVs and Indels analysis, mutations in MUC4, MUC16, ARID1B, BAZ1A, BRCA2, CTNND1 and NCOA2 were the most informative. However, functional annotations of the genes predicted MUC16 as a potential driver gene in our patient with others being either moderately tolerated or benign. Comparison with TCGA data showed that MUC16 had modest mutation frequency in other stage III patients regardless of their age. Although MUC16 mutations is often associated with ovarian cancer (21), emerging studies have linked mutations in the gene with other malignant conditions, including colon cancer (22,23). Further studies are however required to gain full insight into its oncogenic roles. A lack of correlation between MUC16 mutation and survival outcomes in different age groups of TCGA stage III patients may indicate these mutations have no significant influence on the patients’ prognosis. Also, we found no correlation between the age of the patients and their overall survival. Reports on the influence of age on the prognosis of metastatic CRC has not been consistent. Lieu et al. (4) asserted that age was a significant predictor of overall survival in metastatic CRC (stages III and IV) with the younger and older patients showing worse survival than patients of middle age while Schellerer et al. (24) presented opposing results. Inclusion of greater number of patients of particular age group, evaluation of age as a continuous variable, use of older databases among others were cited as probable reasons for the disparity (4). We developed a broad overview of copy number variations in our patient. Totally, we identified 35 copy number alternations including 34 LOH and 1 duplication. TCGA database query of these genes in conventional stage III CRC patients showed they had deep deletions and amplification events. LOH of one gene or another is thought to be relatively common in cancer of all types, however, it is particularly significant in individuals who have inherited a predisposition for cancer suggesting the high possibility of genetic predisposition in our patient (25). Structural variation analysis identified mutations in BAGE2, a B Melanoma Antigen Family Member 2, which to the best of our knowledge has only been reported in CRC patients from the Han Chinese nationality (26). It remains to be determined whether this gene could be a biomarker or a precision therapy target in the population. The disparity observed between our mutation data and that of TCGA demonstrate the highly genetically heterogeneous nature of same stage CRC and this could be due to the differences in the anatomical pathology of CRC as the disease affects different regions of the digestive tract (27). Moreover, genes that drive tumor progression in different regions may be dissimilar, and most CRC studies do not pay specific attention to separating these regions (28). Notably, the use of WGS technique and multiple variant calling tools are the strengths of our study. WGS approach provides a complete coverage of the coding and noncoding regions, enhancing a comprehensive assessment of the patient genome. It also offers a more robust determination of copy number variations, rearrangements and other structural variations due to the longer reads length (29). The use of multiple tools for variant calling facilitates a more accurate and consistent variant identification, providing clinical-grade variant information for genomic medicine (30). However, our study is not without its limitations. Although WGS gave a comprehensive overview of the various alterations in the cancer patient genome, the differential expression profiles of the mutated genes could not be evaluated as we had no transcriptomics data. In conclusion, whole-genome profiling and comparison of our data with TCGA database produced a genomic mutation profile of the patient and offered a valuable resource for further study into the mutations that characterize long-term surviving stage III CRC which may be useful to design clinical therapy for personalized medicine.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.03.55). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of the First Affiliated Hospital of Anhui Medical University (No. PJ-20190110) and is in accordance with the Helsinki Declaration as revised in 2013. Written informed consent was obtained from the patient for publication of this Case report and any accompanying images.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gonzalez-Perez A, Lopez-Bigas N. Functional impact bias reveals cancer drivers. Nucleic Acids Res 2012;40:e169. [Crossref] [PubMed]
- Kamps R, Brandão RD, Bosch BJ, et al. Next-generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci 2017; [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Lieu CH, Renfro LA, de Gramont A, et al. Association of age with survival in patients with metastatic colorectal cancer: analysis from the ARCAD Clinical Trials Program. J Clin Oncol 2014;32:2975-84. [Crossref] [PubMed]
- Chagpar R, Xing Y, Chiang YJ, et al. Adherence to stage-specific treatment guidelines for patients with colon cancer. J Clin Oncol 2012;30:972-9. [Crossref] [PubMed]
- Edge SB, Sobin LH, Page DL, et al. Re: Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst 2005;97:463-4; author reply 464-5.
- Nguyen HT, Duong HQ. The molecular characteristics of colorectal cancer: implications for diagnosis and therapy. Oncol Lett 2018;16:9-18. [PubMed]
- Cui Y, Kong X, Lin F, et al. Efficacy of Huaier granules combined with XELOX regimen in the treatment of stage IV colorectal cancer. China Modern Drug Application 2016;10:11-3.
- Qian J, Zhang L, Jia J, et al. Clinical observation of Huaier granules combined with FOLFOX4 regimen for colorectal cancer. Chinese Journal of General and Basic Medicine 2012;19:429-32.
- Hai Y, Zheng Y, Zhuang Y, et al. Preliminary clinical study of Huaier granule combined with chemotherapy for advanced colorectal cancer. Journal of Pharmacoepidemiology 2012;21:53-5.
- Poplin R, Ruano-Rubio V, DePristo MA, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. BioRxiv 2018;201178: [Crossref]
- Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213-9. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [Crossref] [PubMed]
- McLaren W, Gil L, Hunt SE, et al. The ensembl variant effect predictor. Genome Biol 2016;17:122. [Crossref] [PubMed]
- Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44:e131. [Crossref] [PubMed]
- Team RC. R: A language and environment for statistical computing. 2013.
- Dong C, Wei P, Jian X, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet 2015;24:2125-37. [Crossref] [PubMed]
- Gallego CJ, Shirts BH, Bennette CS, et al. Next-generation sequencing panels for the diagnosis of colorectal cancer and polyposis syndromes: a cost-effectiveness analysis. J Clin Oncol 2015;33:2084-91. [Crossref] [PubMed]
- Susswein LR, Marshall ML, Nusbaum R, et al. Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genet Med 2016;18:823-32. [Crossref] [PubMed]
- Gong R, He Y, Liu XY, et al. Mutation spectrum of germline cancer susceptibility genes among unselected Chinese colorectal cancer patients. Cancer Manag Res 2019;11:3721-39. [Crossref] [PubMed]
- Felder M, Kapur A, Gonzalez-Bosquet J, et al. MUC16 (CA125): tumor biomarker to cancer therapy, a work in progress. Mol Cancer 2014;13:129. [Crossref] [PubMed]
- Haridas D, Ponnusamy MP, Chugh S, et al. MUC16: molecular analysis and its functional implications in benign and malignant conditions. FASEB J 2014;28:4183-99. [Crossref] [PubMed]
- Björkman K, Mustonen H, Kaprio T, et al. Mucin 16 and kallikrein 13 as potential prognostic factors in colon cancer: results of an oncological 92-multiplex immunoassay. Tumour Biol 2019;41:1010428319860728. [Crossref] [PubMed]
- Schellerer VS, Merkel S, Schumann SC, et al. Despite aggressive histopathology survival is not impaired in young patients with colorectal cancer: CRC in patients under 50 years of age. Int J Colorectal Dis 2012;27:71-9. [Crossref] [PubMed]
- Ryland GL, Doyle MA, Goode D, et al. Loss of heterozygosity: what is it good for? BMC Med Genomics 2015;8:45. [Crossref] [PubMed]
- Teng H, Gao R, Qin N, et al. Identification of recurrent and novel mutations by whole-genome sequencing of colorectal tumors from the Han population in Shanghai, eastern China. Mol Med Rep 2018;18:5361-70. [PubMed]
- Linnekamp JF, Wang X, Medema JP, et al. Colorectal cancer heterogeneity and targeted therapy: a case for molecular disease subtypes. Cancer Res 2015;75:245-9. [Crossref] [PubMed]
- Bijlsma MF, Sadanandam A, Tan P, et al. Molecular subtypes in cancers of the gastrointestinal tract. Nat Rev Gastroenterol Hepatol 2017;14:333-42. [Crossref] [PubMed]
- Khromykh A, Solomon BD. The benefits of whole-genome sequencing now and in the future. Mol Syndromol 2015;6:108-9. [Crossref] [PubMed]
- Hwang S, Kim E, Lee I, et al. Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci Rep 2015;5:17875. [Crossref] [PubMed]