
Effects of autophagy-associated genes on the prognosis for lung adenocarcinoma
Introduction
Nowadays, there are over 200,000 lung cancer cases among cancer diseases in 2019 worldwide (1). As the leading cause of death among various cancers (2), the total lung cancer deaths exceed the sum of those of the following three most prevalent cancers, namely, colon, breast and pancreatic cancers (1). The prognosis for lung cancer is relatively poor due to its hidden symptoms at the early disease stage (3). Thus, more attention should be paid to develop the early screening approaches and the novel treatments for lung cancer (2).
As a catabolic process, autophagy maintains the cellular homeostasis through the lysosomes-recycling intracellular components (4,5). As verified by previous studies, autophagy plays various roles, including the neutral, tumor-suppressive, or tumor promoting activities (6-8). At the early stage of cancer, protein and organelles in tumor cells are degraded by autophagy, which relieves the chromosomal instability and cell injury, thus suppressing tumor development (9-11). Nonetheless, autophagy facilitates the survival of cancer cells in the presence of stress conditions when cancer is formed, therefore promoting tumor development (12,13). Besides, those autophagy effects on cancer are examined through the immune system, microenvironmental stress, and nutrient availability (14).
Several studies report that certain scoring systems can be developed, including the immune score, stromal score or DNA damage-repair, to assess the biological processes of tumors (15-17), which reminds us that a scoring system of autophagy may also be built in various tumors (18,19). Autophagy inhibition and stimulation are reported as the treatments for autophagy, and they are also the crucial parts in patient assessment (14,20-22). Furthermore, numerous studies also demonstrate that, the activation of autophagy is remarkably related to chemo-resistance, cancer dormancy and stem cell survival (23).
Previous study has reported the relationship of autophagy with the non-small cell lung cancer (NSCLC) (3). Nonetheless, few studies use the large-scale expression patterns for assessing the autophagy effect on the prognosis for lung adenocarcinoma. This study aimed to build a novel scoring system based on the screened crucial differentially expressed autophagy-related genes (DEARGs), which might serve as a perspective tool in assessing autophagy.
Methods
Gene set related to autophagy
Genes involved in the autophagy process were identified using the human autophagy database (Human Autophagy Database, http://autophag.lu/clustring/index.html).
Patient samples
Gene expression profiles and clinical data of lung adenocarcinoma patients were retrieved from The Cancer Genome Atlas (TCGA) database, and a total of 594 specimens were collected based on TCGA microarray as the cohort.
Processes
Principal component analysis (PCA) was carried out using the R program, to investigate those different expression profiles of genes in the enrolled specimens. The receiver-operating characteristics (ROC) curves of R package were plotted using the survival ROC of R package to assess the survival specificity and sensitivity. Then, the values of area under the ROC curve (AUC) were determined based on those plotted ROC curves. In addition, the online database Kaplan-Meier Plotter (www.Kmplot.com) was applied in verifying the accuracy of our results.
Analyses of functional enrichments
For the differentiated expressed genes (DEGs), the R 3.6.1 was utilized to analyze their functional enrichment for confirming the three categories of Gene Ontology (GO), including biological processes (BP), molecular functions (MF), and cellular components (CC).
Statistical analyses
Multivariate and univariate Cox regression analyses were used for assessing the associations among DEARGs, the risk score value (established by DEARGs) and other clinical baseline characteristics for prognosis prediction. According to the risk score, patients with lung adenocarcinoma were classified as low or high-risk group based on the median risk score. Survival analyses were regarded as the crucial results; and patient prognosis [overall survival (OS)] was compared among various ranges of score using log-rank test and Kaplan-Meier analysis.
The time-dependent ROC curves were plotted to calculate the predictive ability of our risk model. Bioinformatics analysis was carried out using the R software (version 3.6.1). P value of <0.05 was considered as statistically significant. Differences in clinical baseline characteristics between low-risk and high-risk groups were examined through χ2 test or Student’s t-test.
Results
DEARGs within lung adenocarcinoma tissues
Expression of 232 ARGs was analyzed in 535 lung adenocarcinoma and 59 normal lung tissue samples using Wilcoxon signed-rank test, and 30 differentially expressed ARGs (DEARGs) were found. The results showed that 18 of the included DEARGs were up-regulated, and 12 were down-regulated (|log2FC| >1, FDR <0.05, Figure 1).
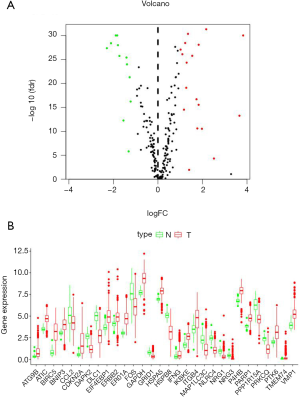
Prognostic factors of DEARGs in patients with lung adenocarcinoma
A total of 31 DEARGs expression within lung adenocarcinoma cases was analyzed through univariate Cox regression analysis for identifying the related DEARGs for prognosis prediction (Figure 2). According to our results, 11 genes (ITGB4, DLC1, HSPB8, GAPDH, NLRC4, BIRC5, ERO1A, DAPK2, and ATIC) were related to the survival of lung adenocarcinoma (Figure 2B). Multivariate Cox proportional hazard regression analysis was also carried out to identify the best model in predicting the patient prognosis. Based on the results, NLRC4 (HR 0.63, 95% CI: 0.43–0.90), ITGB4 (HR 1.14, 95% CI: 1.02–1.28), ERO1A (HR 1.39, 95% CI: 1.14–1.70) and BIRC5 (HR 1.14, 95% CI: 0.98–1.31) were found to be the risk factors that independently predicted survival. The risk score was calculated according to the following formula, risk score = (0.1331 × ITGB4 expression) − (0.4698 × NLRC4 expression) + (0.1270 × BIRC5 expression) + (0.3314 × ERO1A expression). The high-risk DEARGs were related to poor prognosis; on the contrary, the low-risk ones were markedly associated with superior prognosis.
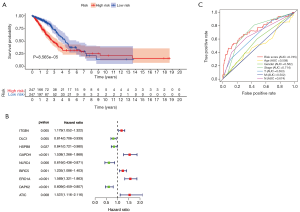
A higher DEARG score was related to the worse OS
Table 1 presents the baseline features of all the included cases. As suggested by our log-rank test results obtained using the Kaplan-Meier survival curve, compared with low-risk cases, the high-risk cases showed poor prognosis (Figure 2A). Therefore, the ROC curves were plotted to determine the effect of the as-constructed signature on patient OS. Based on our results, the AUC value for those prognostic models to predict OS was 0.735. To better predict the prognosis for lung adenocarcinoma patients, baseline characteristics (including age, gender, Tumor, Node, and Metastasis stage in all patients) were used in ROC curve analysis. Then, their distributions were ranked and analyzed, which revealed that stage (AUC 0.714), node stage (AUC 0.674), tumor stage (AUC 0.653), gender (AUC 0.562), age (AUC 0.508), and metastasis stage (AUC 0.502) were followed by the risk score (Figure 2C).
Table 1
| Characteristics | Total (n=522) | Percentages (%) |
|---|---|---|
| Age, years | ||
| >65 | 262 | 50.19 |
| ≤65 | 241 | 46.17 |
| Unknown | 19 | 3.64 |
| Gender | ||
| Female | 280 | 53.64 |
| Male | 242 | 46.36 |
| Stage | ||
| Stage I | 279 | 53.45 |
| Stage II | 124 | 23.75 |
| Stage III | 85 | 16.28 |
| Stage IV | 26 | 4.98 |
| Unknown | 8 | 1.53 |
| T | ||
| T1 | 172 | 32.95 |
| T2 | 281 | 53.83 |
| T3 | 47 | 9.00 |
| T4 | 19 | 3.64 |
| Unknown | 3 | 0.57 |
| M | ||
| M0 | 353 | 67.62 |
| M1 | 25 | 4.79 |
| Unknown | 144 | 27.59 |
| N | ||
| N0 | 335 | 64.18 |
| N1 | 98 | 18.77 |
| N2 | 75 | 14.37 |
| N3 | 2 | 0.38 |
| Unknown | 12 | 2.30 |
| Survival rate | ||
| Survival (n=1) | 167 | 31.99 |
| Death (n=0) | 355 | 68.01 |
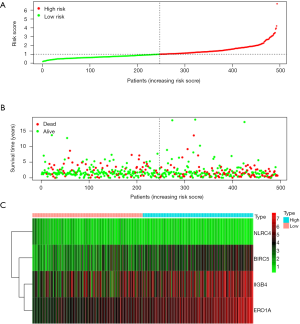
As shown in the figure, high-risk cases in OS group had up-regulated BIRC5, ITGB4, and ERO1A, whereas down-regulated NLRC4. Figure 3 displays the distributions of risk gene expression, survival status and risk score among these cases.
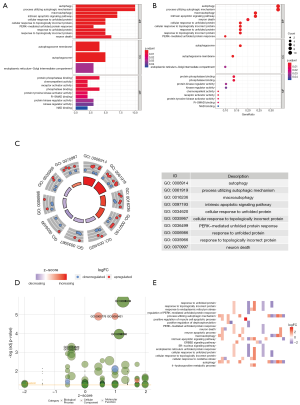
GO functional enrichment analysis
Therefore, functional enrichment analysis was carried out on these genes. According to those 10 most significant GO terms with regard to MF, CC and BP categories; the above-mentioned DEGs were potentially associated with immune cell regulation, ECM, cytokine activation and binding of receptors (Figure 4).
Multivariate Cox regression analysis on risk score as well as baseline clinical features
The results showed that the tumor stage (HR 1.861, 95% CI: 1.159–2.990), and risk score (HR 2.052, 95% CI: 1.621–2.597) were the independent risk factors of survival (Figure 5).
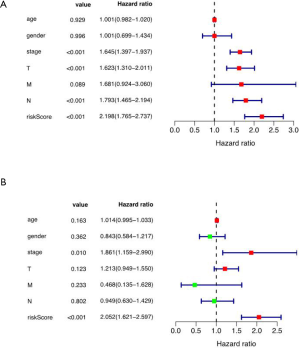
Associations of risk signature and DEARGs with clinical features (including gender, age, stage, T, N, M stage)
As demonstrated by our results, BIRC5, ERO1A, NLRC4 and risk score were related to the survival status and stage (Figure 6).
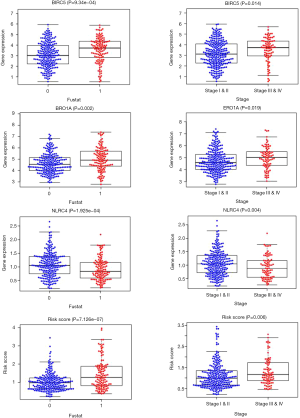
The Kaplan-Meier Plot dataset was used for result verification
The results showed that these four genes were related to the poor OS and progression-free survival (PFS); however, difference in the post-progression survival (PPS) was not statistically significant (Figure 7).
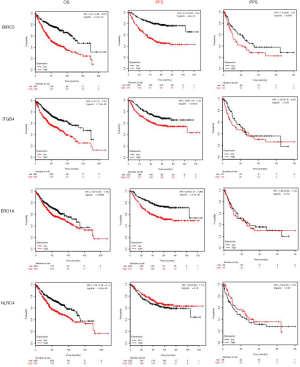
Discussion
NSCLC is generally classified as three principle histological subtypes, including lung adenocarcinoma (40–70%), large-cell carcinoma (10–15%), and squamous-cell carcinoma (20–30%) (24). This study aimed to find out a more helpful approach to predict the prognosis for lung adenocarcinoma.
Under physiological situation, autophagy functions as an intracellular quality control system to maintain the homeostasis; besides, it is mainly responsible for removing the damaged proteins (25). Previously, Mathew et al. (26) indicated that, the loss of autophagy within pancreas and liver resulted in oxidative stress, inflammation as well as chronic tissue damage, which revealed the benefits of autophagy in tumor initiation. Autophagy is the new therapeutic approach, and its process in tumor has long been proposed. Nevertheless, according to the results by Sui et al. (27), autophagy probably affected patient responses to chemoradiotherapy, which worked as an assisting tool.
Firstly, this study found the different expression of autophagy between lung adenocarcinoma and normal lung tissues, which might explain the role of autophagy. However, no consensus is reached for the controversial functions of autophagy so far (28); therefore, it may be helpful to develop a useful autophagy-related risk score to assist the clinical workers in better applying the autophagy-targeted therapy for lung adenocarcinoma patients. Obviously, compared with other classical clinical indicators, the risk score calculated by the DEARGs score was superior.
In previous studies, Liu et al. indicated that the processes of autophagy and apoptosis were crucial in the pathophysiology of NSCLC (3). First of all, plenty of evidence demonstrates that autophagy assists in the migration and invasion of lung cancer cells by toll-like receptors (TLR4)- triggered by TL3 (29). Moreover, several studies report that various substances are beneficial for lung cancer patients, such as Marsdenia tenacissima (30), Juglanin (31), SBI0206965 (32), di-limonene (33), CK1α (34) and miR-138 (35). Lastly, some drugs not only play certain roles in the autophagy, but also target some other antigens [including CD47 (36) and ULK1(37)], thus rendering them with the synergistic antitumor effects.
Among the independent risk genes included in this study, we are especially interested in BIRC5 and ERO1A. Among them, the high BIRC5 level indicates cancer development of and dismal prognosis for lung adenocarcinoma (38). Knocking down BIRC5 induces the apoptosis of cancer cells, which is thereby considered to be an underlying target of AML treatment (39). For ERO1A, Takei et al. (40) indicated that it was over-expressed in various cancer types, and its expression acted as a novel factor related to poor cancer prognosis. Additionally, Tanaka et al. (41) also showed that, ERO1A played an important part in the formation of disulfide bond under the combined action of the programmed cell death-1 (PDI). Moreover, ERO1A regulates MHC class I expression on cell surface and redox status, thus resulting in changes in CD8+ T cells susceptibility (42).
Conclusions
All in all, DEARGs are used to successfully establish a novel scoring system in this study, which accurately assesses the outcomes for patients with lung adenocarcinoma. The included DEARGs are confirmed by the online dataset called Kaplan-Meier Plotter (www.Kmplot.com). According to our results, DEARGs are regarded as the treatment targets and prognostic biomarkers within lung adenocarcinoma. Moreover, our results should be verified in additional studies.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.02.07). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The data comes from database, so ethical reviews are exempt. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol 2016;893:1-19. [Crossref] [PubMed]
- Liu G, Pei F, Yang F, et al. Role of Autophagy and Apoptosis in Non-Small-Cell Lung Cancer. Int J Mol Sci 2017; [Crossref] [PubMed]
- Bouhamdani N, Comeau D, Cormier K, et al. STF-62247 accumulates in lysosomes and blocks late stages of autophagy to selectively target von Hippel-Lindau-inactivated cells. Am J Physiol Cell Physiol 2019;316:C605-20. [Crossref] [PubMed]
- Cohignac V, Landry MJ, Ridoux A, et al. Carbon nanotubes, but not spherical nanoparticles, block autophagy by a shape-related targeting of lysosomes in murine macrophages. Autophagy 2018;14:1323-34. [Crossref] [PubMed]
- Shi L, Yan H, An S, et al. SIRT5-mediated deacetylation of LDHB promotes autophagy and tumorigenesis in colorectal cancer. Mol Oncol 2019;13:358-75. [Crossref] [PubMed]
- Rao S, Tortola L, Perlot T, et al. A dual role for autophagy in a murine model of lung cancer. Nat Commun 2014;5:3056. [Crossref] [PubMed]
- Karsli-Uzunbas G, Guo JY, Price S, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov 2014;4:914-27. [Crossref] [PubMed]
- Cao Q, Bai P. Role of Autophagy in Renal Cancer. J Cancer 2019;10:2501-9. [Crossref] [PubMed]
- New M, Tooze S. The Role of Autophagy in Pancreatic Cancer-Recent Advances. Biology (Basel) 2019; [Crossref] [PubMed]
- Cotzomi-Ortega I, Aguilar-Alonso P, Reyes-Leyva J, et al. Autophagy and Its Role in Protein Secretion: Implications for Cancer Therapy. Mediators Inflamm 2018;2018:4231591.
- Folkerts H, Hilgendorf S, Vellenga E, et al. The multifaceted role of autophagy in cancer and the microenvironment. Med Res Rev 2019;39:517-60. [Crossref] [PubMed]
- Cao Y, Luo Y, Zou J, et al. Autophagy and its role in gastric cancer. Clin Chim Acta 2019;489:10-20. [Crossref] [PubMed]
- Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev 2016;30:1913-30. [Crossref] [PubMed]
- Evans JR, Zhao SG, Chang SL, et al. Patient-Level DNA Damage and Repair Pathway Profiles and Prognosis After Prostatectomy for High-Risk Prostate Cancer. JAMA Oncol 2016;2:471-80. [Crossref] [PubMed]
- Mlecnik B, Bindea G, Angell HK, et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity 2016;44:698-711. [Crossref] [PubMed]
- Mlecnik B, Bindea G, Kirilovsky A, et al. The tumor microenvironment and Immunoscore are critical determinants of dissemination to distant metastasis. Sci Transl Med 2016;8:327ra26. [Crossref] [PubMed]
- Wan B, Liu B, Yu G, et al. Differentially expressed autophagy-related genes are potential prognostic and diagnostic biomarkers in clear-cell renal cell carcinoma. Aging (Albany NY) 2019;11:9025-42. [Crossref] [PubMed]
- Zhou Z, Mo S, Dai W, et al. Development and Validation of an Autophagy Score Signature for the Prediction of Post-operative Survival in Colorectal Cancer. Front Oncol 2019;9:878. [Crossref] [PubMed]
- Tyutyunyk-Massey L, Gewirtz DA. Roles of autophagy in breast cancer treatment: Target, bystander or benefactor. Semin Cancer Biol 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer 2017;17:528-42. [Crossref] [PubMed]
- Towers CG, Thorburn A. Therapeutic Targeting of Autophagy. EBioMedicine 2016;14:15-23. [Crossref] [PubMed]
- Galluzzi L, Pietrocola F, Levine B, et al. Metabolic control of autophagy. Cell 2014;159:1263-76. [Crossref] [PubMed]
- Neal RD, Hamilton W, Rogers TK. Lung cancer. Bmj 2014;349:g6560. [Crossref] [PubMed]
- Marino G, Niso-Santano M, Baehrecke EH, et al. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 2014;15:81-94. [Crossref] [PubMed]
- Mathew R, Karantza-Wadsworth V, White E. Assessing metabolic stress and autophagy status in epithelial tumors. Methods Enzymol 2009;453:53-81. [Crossref] [PubMed]
- Sui X, Chen R, Wang Z, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis 2013;4:e838. [Crossref] [PubMed]
- An Y, Bi F, You Y, et al. Development of a Novel Autophagy-related Prognostic Signature for Serous Ovarian Cancer. J Cancer 2018;9:4058-71. [Crossref] [PubMed]
- Zhan Z, Xie X, Cao H, et al. Autophagy facilitates TLR4- and TLR3-triggered migration and invasion of lung cancer cells through the promotion of TRAF6 ubiquitination. Autophagy 2014;10:257-68. [Crossref] [PubMed]
- Jiao YN, Wu LN, Xue D, et al. Marsdenia tenacissima extract induces apoptosis and suppresses autophagy through ERK activation in lung cancer cells. Cancer Cell Int 2018;18:149. [Crossref] [PubMed]
- Chen L, Xiong YQ, Xu J, et al. Juglanin inhibits lung cancer by regulation of apoptosis, ROS and autophagy induction. Oncotarget 2017;8:93878-98. [PubMed]
- Tang F, Hu P, Yang Z, et al. SBI0206965, a novel inhibitor of Ulk1, suppresses non-small cell lung cancer cell growth by modulating both autophagy and apoptosis pathways. Oncol Rep 2017;37:3449-58. [Crossref] [PubMed]
- Yu X, Lin H, Wang Y, et al. d-limonene exhibits antitumor activity by inducing autophagy and apoptosis in lung cancer. Onco Targets Ther 2018;11:1833-47. [Crossref] [PubMed]
- Cai J, Li R, Xu X, et al. CK1alpha suppresses lung tumour growth by stabilizing PTEN and inducing autophagy. Nat Cell Biol 2018;20:465-78. [Crossref] [PubMed]
- Ye Z, Fang B, Pan J, et al. miR-138 suppresses the proliferation, metastasis and autophagy of non-small cell lung cancer by targeting Sirt1. Oncol Rep 2017;37:3244-52. [Crossref] [PubMed]
- Zhang X, Fan J, Wang S, et al. Targeting CD47 and Autophagy Elicited Enhanced Antitumor Effects in Non-Small Cell Lung Cancer. Cancer Immunol Res 2017;5:363-75. [Crossref] [PubMed]
- Martin KR, Celano SL, Solitro AR, et al. A Potent and Selective ULK1 Inhibitor Suppresses Autophagy and Sensitizes Cancer Cells to Nutrient Stress. iScience 2018;8:74-84.
- Cao Y, Zhu W, Chen W, et al. Prognostic Value of BIRC5 in Lung Adenocarcinoma Lacking EGFR, KRAS, and ALK Mutations by Integrated Bioinformatics Analysis. Dis Markers 2019;2019:5451290.
- Narimani M, Sharifi M, Hakhamaneshi MS, et al. BIRC5 Gene Disruption via CRISPR/Cas9n Platform Suppress Acute Myelocytic Leukemia Progression. Iran Biomed J 2019;23:369-78. [Crossref] [PubMed]
- Takei N, Yoneda A, Sakai-Sawada K, et al. Hypoxia-inducible ERO1alpha promotes cancer progression through modulation of integrin-beta1 modification and signalling in HCT116 colorectal cancer cells. Sci Rep 2017;7:9389. [Crossref] [PubMed]
- Tanaka T, Kutomi G, Kajiwara T, et al. Cancer-associated oxidoreductase ERO1-alpha promotes immune escape through up-regulation of PD-L1 in human breast cancer. Oncotarget 2017;8:24706-18. [Crossref] [PubMed]
- Kukita K, Tamura Y, Tanaka T, et al. Cancer-Associated Oxidase ERO1-alpha Regulates the Expression of MHC Class I Molecule via Oxidative Folding. J Immunol 2015;194:4988-96. [Crossref] [PubMed]


