Metallothionein 1M (MT1M) inhibits lung adenocarcinoma cell viability, migration, and expression of cell mobility-related proteins through MDM2/p53/MT1M signaling
Introduction
Lung cancer is still the most significant health problem in the world, accounting for an estimated 11.6% of the 17 million new cancer cases and 18.4% of the 9.5 million cancer deaths in 2018 globally (1). In China, lung cancer is also the most frequently diagnosed malignancy and was the leading cause of cancer death in 2015 (2). To date, the majority of lung cancer patients are diagnosed at advanced stages of the disease, making curable tumor surgery impossible. Non-surgical treatment of non-small cell lung cancer includes chemoradiation therapy and the recently discovered immune and targeting therapy (1,3-5). The latter treatment option can effectively control lung cancer progression and prolong the survival of patients (6,7). Thus, further efforts to identify novel targets and lung cancer pathogenesis could lead to better control of lung cancer clinically.
To this end, metallothionein (MT) 1M belongs to a family of cysteine-rich, low molecular mass proteins localized on the membrane of the Golgi apparatus (6-8) and is involved in metalloregulatory processes. For example, MTs can regulate the cancer cell life cycle, including proliferation and dissemination, due to their high affinity to binding metals such as Zinc and Copper as well as heavy metals (Cadmium) to protect cells from metal toxicity (9-14). Thus, MTs play a role in cancer development and progression (15,16). The metallothionein 1M (MT1M) functions and underlying molecular mechanisms have been studied in various human cancers. For example, methylation of the MT1M promoter occurs in hepatocellular carcinoma (HCC) (17) and MT1M inhibition is associated with poor HCC prognosis (18). However, MT1M overexpression inhibits HCC cell viability and induces the cells to undergo apoptosis in vitro and in nude mouse xenograft (19,20). In papillary thyroid cancer tissues, MT1M expression is reduced (21). MT1M overexpression inhibits papillary thyroid cancer cell proliferation, colony formation, migration, and invasion (21). However, there have only been a few studies on MT1M in lung cancer. A recent study showed that high DNA methylation, including MT1M, is inversely associated with lung squamous cell carcinoma (SCC), idiopathic pulmonary fibrosis, and poorer SCC prognosis vs. low DNA methylation, indicating that MT1M has an opposite effect in lung SCC compared with its effect in HCC and thyroid cancer (22). Furthermore, our unpublished study on the mRNA array dataset for interstitial lung disease (GSE81292) revealed that MT1M expression is downregulated in lung cancer.
In this study, we investigated the effects of MT1M overexpression and mouse double minute 2 homolog (MDM2) knockdown on the regulation of non-small cell lung cancer A549 cell viability, migration, and gene expression in vitro and then explored the underlying molecular events. We first stably infected A549 cells with lentivirus carrying MT1M cDNA or transiently transfected MDM2 siRNA into the cells and then treated them with a p53 inhibitor. Afterward, we assessed the changes in cell viability, wound healing, and Transwell migration and used a Luciferase reporter assay to evaluate p53 binding to the MT1M promoter. We expect the results to provide novel information regarding MT1M in the regulation of lung cancer cell malignant behaviors to aid in a future strategy for targeting therapy for lung cancer.
Methods
Cell line and culture
A human NSCLC A549 cell line was originally purchased from American Type Culture Collection (Manassas, VA, USA) and cryopreserved in our laboratory. In this study, A549 cells were cultured in an F-12K medium (ATCC) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) in a humidified incubator with 5% CO2 at 37 °C.
Construction of MT1M expression plasmids, lentivirus production, MDM2 shRNA, and cell infection
In this study, we constructed a vector carrying the MT1M coding sequences using the pLVX-puro plasmid (Youbio, Changsha, China) according to the manufacturer’s cloning protocols. After DNA sequencing confirmation, the vector carrying MT1M coding sequences and the vector-only control were used to transfect HEK-293T cells for lentivirus production. Afterward, A549 cells were grown and infected with these lentiviruses at a multiplicity of infection of 20 in 8 µg/mL polybrene (Sigma, St. Louis, MO, USA) for 48 h and then observed under an inverted microscope (Olympus, Tokyo, Japan). Next, the cell culture medium was replaced with medium containing 1.0 µg/mL puromycin to establish a stable cell population. Meanwhile, a set of infected cells was treated with the p53 inhibitor (Selleck, Shanghai, China) at a final concentration of 10 µM or transiently transfected with MDM2 siRNA using Lipofectamine 2000 Transfection Reagent (Thermo, Waltham, MA USA) for 48 h according to the manufacturer’s instructions then subjected to different analyses (see below for details). The MDM2 siRNA with the targeting sequences of 5'-UUUAUAAUCACAUUUAUGCUA-3' was synthesized and obtained from GenePharma (Shanghai, China). The negative control siRNA was also purchased from GenePharma.
Reverse transcription and real-time quantitative PCR (RT-qPCR)
Total cellular RNA was isolated from cells using the TRIzol reagent (GENEray, Shanghai, China) according to the manufacturer’s instructions and the concentrations of the RNA samples were determined using NanoDrop (Thermo). Next, the samples were reverse-transcribed using a cDNA GENEray kit (GENEray, Shanghai, China) according to the manufacturer’s protocol. Afterward, qPCR was amplified using the cDNA samples with the Geneseed® qPCR SYBR® Green Master Mix (TaKaRa, Dalian, China) in an ABI 7500 machine (ABI, Foster city, CA, USA) according to the manufacturer’s instructions. The qPCR conditions were set to an initial 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 60 s. The primers used were MT1M, 5'-GCAAGTGCAAAGAGTGCAAATG-3', and 5'-CTGCAGTTCTCCAACGTCC-3' (which generates a 118-bp qPCR product); MDM2, 5'-ACGACAAAGAAAACGCCACA-3' and 5'-GTAACTTGATATACACCAGCATCAA-3' (to obtain a 162-bp product); and β-actin, 5'-AACTGGGACGACATGGAGAAAA-3', and 5'-GGATAGCACAGCCTGGATAGCA-3' (to obtain a 192-bp qPCR product). The relative value of MT1M and MDM2 mRNA expression was compared to the internal control of β-actin and expressed as 2-(Ct-Cc) (Ct and Cc were the mean threshold cycle differences after normalizing to β-actin).
Western blot
The cultured cells were lysed with the sodium dodecyl sulfate (SDS) lysis buffer (Beyotime, Shanghai, China) on ice for 10 min and centrifuged at 4 °C for 15 min at the maximal speed. The supernatants were collected into new Eppendorf tubes for assessment of the concentration using the enhanced bicinchoninic acid (BCA) protein assay kit (Beyotime) and denaturalized in a water bath at 100 °C for 5 min. Equal amounts of the protein samples were separated in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA) followed by Western blotting. The membranes were incubated with anti-MDM2 (Abcam, Cambridge, MA, USA), anti-MT1M (Abbexa, Cambridge, UK), anti-p53 (Abcam), anti-phosphorylated p53 (Abcam), anti-MMP14 (Abcam), anti-MMP9 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-MMP2 (Abcam) or anti-β-actin (MultiScience, Hangzhou, Zhejiang, China) antibody and then with the corresponding secondary antibodies according to the manufacturer’s Western blot protocols. The relative protein levels were detected with a peroxide LumiGLO reagent (Cell Signaling Technology, Danvers, MA, USA) and quantified according to the ratio of the gray value to the corresponding β-actin levels. The experiments were repeated three times independently.
Co-immunoprecipitation (Co-IP)-Western blot
Cells were first lysed using the cell lysis buffer (Beyotime) and protein concentration was measured using the Enhanced BCA Protein Assay Kit (Beyotime). One milligram of each protein sample was incubated with anti-MDM2 antibody (Abcam) at 4 °C overnight. On the next day, the antibody-cell lysis mixture was incubated with Dynabeads™ M-280 Tosylactivated (Thermo) at 4 °C for 4 h and the precipitated protein samples were separated and washed through centrifugation. The collected protein samples were subjected to Western blot with anti-MDM2 or anti-p53 antibody.
Cell viability cell-counting kit 8 (CCK-8) assay
After exposure to various treatments and growth conditions, cells were re-seeded into 96-well plates with 2,000 cells per well in 100 µL culture medium and each condition was quadrupled. The cells were grown for up to 24–96 h and at the end of each experiment, the cell culture medium was supplemented with 10 µL CCK8 solution (MultiScience). The cells were further grown for 1 h and the optical density (OD) value was measured using a microplate reader (Microdevices, San Jose, CA, USA) at 450 nm. The cell viability rate was summarized as the mean ± SD (n=4). The experiments were repeated at least three times.
Wound-healing assay
After exposure to various treatments and growth conditions, cells were re-seeded into six-well culture plates in triplicate and cultured overnight to reach approximately 90–95% confluency. Wounds were made across the cell culture monolayer using a 200-µL pipette tip and the cell debris was washed with ice-cold phosphate buffered saline (PBS). The cells were further cultured at 37 °C for 24 h. The wound healing distance was measured immediately (0 h) and 24 h after the scratch was made using photos taken under an inverted microscope (Olympus). The percentage of the scratch remaining was calculated as: (measurement at 24 h/measurement at 0 h) ×100% and the percentage of wound closure was calculated as: 100% – percentage of wound remaining as mean ± SD.
Migration assay
After exposure to various treatments and growth conditions, the cells were suspended in F-12K medium without FBS supplementation and seeded into the upper chamber of Millicell hanging cell culture inserts (Merck Millipore, Darmstadt, Germany). A complete culture medium containing 20% FBS was added to the lower chambers of the 24-well culture plate and the cells were cultured for 24 h. The cells that migrated into the other side of the filters were fixed with 70% methanol and stained with 0.5% crystal violet solution. The images of the migrated cells from five randomly selected fields (×200) were photographed under an inverted microscope (Olympus) and counted. The data were expressed as mean ± SD (n=3).
Bioinformatic analysis of MT1M-related genes and Luciferase reporter assay
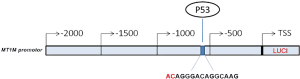
To explore the MT1M protein signaling, we first performed bioinformatic analysis using Jaspar2018 online tools (http://jaspar.genereg.net/) and found that p53 could regulate MT1M expression through p53 binding to the MT1M promoter region (see Figure S1).
MT1M promoter sequences with a length of 2,000 bp (see supplementary material) were PCR-amplified and subcloned into the pGL3-Basic vector using the ClonExpress II One Step Cloning Kit (Vazyme Biotech, Nanjing, China) and designated as the pGL3-MT1M vector. The pGL3-MT1M mutant vector was constructed using the Express MultiS Fast Mutagenesis Kit V2 (Vazyme Biotech) and the mutated sites are presented in the Supplemental material.
To further assess the p53-regulating MT1M expression, we constructed luciferase reporter plasmids using wild type or mutated MT1M promoter and performed the luciferase reporter assay. Briefly, HEK-293T cells were plated into a 24-well plate and grown overnight. The cells were co-transfected with 0.2 µg of the pGL3-MT1M promoter or pGL3 vector only along with 1 µg of p53 expression vector or a negative control using Lipofectamine 2000 for 48 h. Afterward, the cells were harvested and the luciferase activity was assayed using a dual luciferase assay system (Promega, Madison, WI, USA). Each transfection was performed in triplicate.
Statistical analysis
The data were summarized as mean ± SD and statistically analyzed with the Student’s t-test for comparison between two groups. P<0.05 was considered statistically significant.
Results
MT1M overexpression inhibits A549 viability and migration
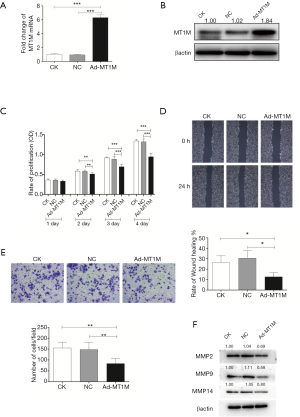
Previous studies reported reduced MT1M expression in thyroid cancer and HCC (19,21,23). Our analysis of differentially expressed mRNA array data (GSE81292) also showed lost MT1M expression in lung adenocarcinoma (LUAD). Thus, in this study, we selected a LUAD A549 cell line to assess the role of MT1M in the regulation of non-small cell lung cancer cell malignant behaviors in vitro. We overexpressed MT1M protein in A549 cells using lentivirus carrying MT1M coding sequences (Figure 1A,B) and our cell viability assay showed that A549 cell viability was significantly reduced compared to the vector-only control (NC)-infected cells (Figure 1C). Our wound healing and Transwell cell migration assays revealed that the migration ability of A549 cells after MT1M overexpression was attenuated (Figure 1D,E). Furthermore, the expression of cell migration-related proteins, including MMP2, MMP9, and MMP14, assayed using Western blot was significantly decreased after MT1M overexpression (Figure 1F). Taken together, MT1M overexpression inhibited A549 cell viability and dissemination, indicating that MT1M could be a tumor suppressor or at least possess anti-tumor activity in non-small cell lung cancer.
p53 inhibition reverses the effects of MT1M overexpression-inhibited A549 cell viability and migration
Next, we explored the underlying molecular events of MT1M anti-lung cancer cell activity through bioinformatic analysis and found that p53 could regulate MT1M expression through p53 binding to the MT1M promoter region. We thus constructed luciferase reporter plasmids using wild type and mutated MT1M promoters and assessed the changes in luciferase activity in HEK-293 cells. Our data showed that luciferase activity dramatically increased in p53 and MT1M promoter co-transfected cells (Figure 2A), indicating that p53 was able to bind to the MT1M promoter to upregulate MT1M expression.
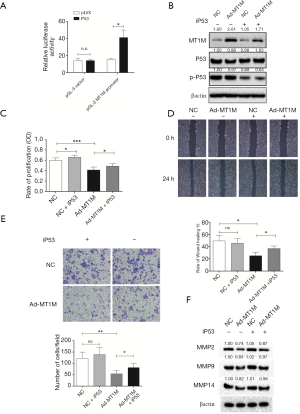
Next, we treated A549 parental and MT1M-overexpressed cells with a p53 inhibitor. Our Western blot data showed that blockage of the p53 activity reduced MT1M expression (Figure 2B), whereas p53 did not affect lentivirus-induced MT1M overexpression (Figure 2B), indicating that p53 can upregulate MT1M expression in A549 cells. Moreover, cell viability and migration assays revealed that the p53 inhibitor was able to mitigate the inhibitory effects of MT1M on A549 cell viability and migration capacity (Figure 2C,D,E). In addition, the expression of cell mobility-related proteins like MMP2, MMP9, and MMP14 also partially recovered after treatment with the p53 inhibitor (Figure 2F).
Knockdown of MDM2 expression reduces A549 cell viability and migration
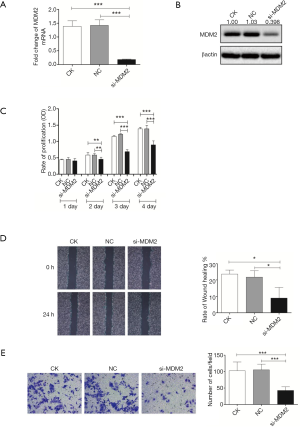
Thus far, we have demonstrated that MT1M anti-tumor activity in non-small cell lung cancer A549 cells in vitro is modulated by p53 activity. We next knocked down MDM2 expression using the MDM2 siRNA to inhibit p53 in cells because MDM2 can downregulate p53 (24) and found reduced levels of MDM2 mRNA and protein in A549 cells (Figure 3). The anti-MDM2 antibody was also able to precipitate p53 in A549 cell lysates (Figure 4A), further providing evidence of their interaction and regulation. Our cell viability and migration assay showed that after MDM2 knockdown, the viability and migration capacity of A549 cells were significantly slower than those of control cells (Figure 3C,D,E). These data suggest that MDM2 plays a role in the reduction of A549 cell viability and dissemination in vitro.
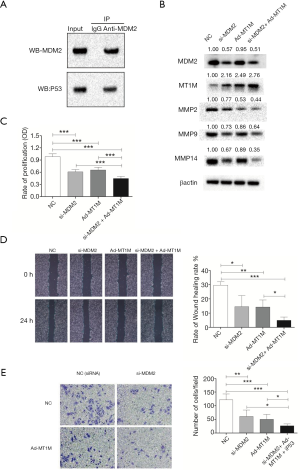
MDM2 knockdown synergizes with the inhibitory effect of MT1M overexpression on A549 cells
We next assessed whether MDM2 knockdown can synergize with the inhibitory effect of MT1M overexpression on A549 cells. The MDM2 siRNA was able to knockdown MDM2 expression and reduce MT1M expression in A549 cells (Figure 4B), indicating that MDM2 regulated MT1M expression. We then assessed the cell viability and migration as well as protein expression and found that MDM2 knockdown synergizes with the inhibitory effect of MT1M overexpression on A549 cell viability and migration as well as protein expression (Figure 4B,C,D,E).
Discussion
In this study, we assessed MDM2-p53-MT1M signaling in the regulation of NSCLC A549 cell viability, migration, and gene expression. Our data revealed that MT1M overexpression was able to reduce A549 cell viability and migration capacity in vitro, and the p53 inhibitor reversed these effects on A549 cells. Furthermore, knockdown of MDM2 expression using MDM2 siRNA vs. the negative control siRNA also reduced A549 cell viability, migration, and protein expression. Thus, MDM2 knockdown had synergistic effects with MT1M overexpression on the suppression of A549 cell viability, migration, and protein expression. Overall, our current study indicates that MDM2 binding to and phosphorylation of p53 protein inactivates the protein to reduce MT1M expression, resulting in A549 cell proliferation and migration.
Indeed, MTs play an important role in the regulation of gene transcription factors. Altered MT expression and functions could lead to the malignant transformation of cells and cancer development (25). However, a previous study showed the differential expression of MTs in various cancers, i.e., upregulation in breast, colon, kidney, liver, melanoma, lung, nasopharynx, ovary, prostate, mouth, salivary gland, testes, thyroid, and urinary bladder cancers, but downregulation in HCC and liver adenocarcinoma (26,27). Another study showed that higher levels of MT expression contributed to chemotherapy resistance (28). In our current study, we revealed that MT1M possessed anti-LUAD activity in vitro, which contrasts with the behavior in lung SCC but is consistent with previous studies on HCC, papillary thyroid cancer, and breast cancer (21,29). Thus, further study is needed to confirm MT1M anti-LUAD activity in vitro and in vivo, although our unpublished analysis of the online microarray data (the GSE81292 dataset) showed reduced MT1M expression in LUADs.
Furthermore, p53 is a tumor suppressor protein in a variety of human cancers, including lung cancer (30). p53 is an important transcription factor that regulates cell activity by manipulating downstream protein expression. p53 activity and levels are tightly regulated in multiple ways, including phosphorylation, acetylation, and ubiquitination (31). p53 functions to regulate the expression of targeting proteins involved in the control of DNA repair, cell cycle progression, apoptosis, and genome stability. For example, p53 expression is maintained at a proper level in normal cells, but is upregulated to determine DNA repair or apoptosis in stressed cells (32). Moreover, MDM2 can bind to p53 protein and inactivate p53 functions and actions by transporting the protein from the nucleus to the cytosol. MDM2 can also act as an ubiquitin ligase to degrade p53 protein (32). In addition, when p53 expression is upregulated, MDM2 expression is also upregulated, which leads to a negative feedback loop to tightly control p53 level in the cells (33). Thus, during periods of cell stress, p53 protein could determine the cells that survive or undergo apoptosis (34). Our current study revealed a novel MDM2-p53 regulation of MT1M expression in A549 cells to inhibit MT1M expression and subsequently suppress MT1M antitumor activity in vitro. However, a previous study of p53 and MT in the apoptosis of shrimp hemocytes in response to hypoxia showed that apoptosis in normoxia was significantly higher in p53-and MT-knocked down animals compared to the level in the negative controls, indicating that high mRNA levels of p53 and MT may not be necessary for the hypoxia response (35). Another previous study demonstrated a direct association of p53 and MTs, but indicated that wild type p53 might be a negative regulator of MT-3 expression in breast cancer cells (36).
However, our current study is just a proof-of-principle and more studies are needed to confirm the role of MDM2-p53-MT1M in the regulation of the fate and survival of patients with LUAD.
Conclusions
In the current study, we investigated the effects of MT1M, p53, and MDM2 on the regulation of A549 cell malignant behaviors in vitro. The results showed that both MTM1 and p53 were able to inhibit A549 cell proliferation and migration, whereas MDM2 had the opposite effect. Future study will confirm the targeting of this novel signaling pathway in the control of LUAD.
Supplementary
MT1M promoter and its mutant sequence cloned into pGL3-Basic vector
MT1M promoter-wild:
AGCATAGGGGATATTTATTTATTTGCACATAAAGGCCTGGAGTGCTCAGCTGTGTATCCTGTTATCCTCATCTACCGCATGAGCATGTGTCCTGTGGTCTAAAGAGAGCCATTCAGTGTCCCCACAAAATTGTTATCAACACTAGGGTTATAAAGCTATGAGGAAGATTCTGTGCTTCATGTCAGATACAGAAGAATGACTGCCATTTCTGTTACCACCTGCACCATCCCTTAGTTAATATTTTACCTTGAAATGATCAATACTTCTTAAACATATTTTTATGAAAAGCAAATTATTTTCCACCTGAGAAAATGAAATCCAATCTTGTCTTACACATGGAAGATAGCCCCCAAATTAATGTGTGTGTGTGTGTGTGTGTATTACACTTATGGATTACTTTTGCCACTGCCCATCAGAGAGCTGGTAGCTTCCTGTTGTTTATTTGAGGGTTTTTGGTTGTTTTTGTTTGTTGAGACAGAGTTTTACCCCTGTCAGCCAGCTGGAGTACAATGGCCCAATCTGCCACCTCTGCCTCCCGGGTTCAAACGATTTTCCTGCCCAAGCCTCCCCAGTAGCTGGGATTACAGGCAACTGCCACCATGCCAGGATAATTTTTGTATTTTTAGTGGAGACAGGGTTTCGCCATGTTAGCCAGGCTGGTCTCCAACTCCTGACCTCAGGTGATTCATCCGCCTCGGCCTTCGTAAGTGCTGGGATTACAGGTGTGGGCCGCGGTGCCCTTCCCCTGTTGTTTAGAAGATTTCCAGCAATAGAGAAGTGCTGACGACATATCTGGACCAAAGGAAGGCTTGATCATGGACACTTTCCTTTAAGAGGCTGGCTGGACTTAAACAAGGGCACAAATTTTTAGCTCTGTGACGACCAGCCAGTTACTTGGACATTTCCTTTGTAGTCCTGTGATGAGCAAAACGGGTTCATAACTAGTATTTCCCTTTAGGATTGCTGTGAGGATTCAAAGAATGAAAGAGATTAAAAAGGGACTGGTGTAGACTCAATGTCATGCAATAGTCAGGTATACTGAAGAGAAACAACTGCATGTCTTGTTTTTTAATTTTATTATTTTAGAGATGCTGACTTGCTATGTGGACTAGGCAGAACTCGAACTCCTGGGCTCAGGTGATCCTCCCGCCTCAGCCTCCTAAGTAGCTGGGACTACAGCTTCCCGCGACCTCACCCATCTTGCTTTTTAATTTAAAGCAGAGTCTGCATATCATCTGAAGTCATCTCTCTTTGGGGACATCCCACAGGTCCAGAACTGCCAGACGGTAGTGGGGTGGCCGGCTAGGCTGTGGGGAGCATGGAGATTTATCTGCAAAGGAGGACCTGGACAAATGTTCCCCCACATCCTCTCAGGCGAAGAGAATGGACGGGAGAGAGAGGCCGACCAGTGTTCCCCGTGTTGCTGTGTACGGAGGAGCCAGTCCGAGGGACCGCGGTGTGGACAGGGACAGGCAAGGCGGGGAAGGAGGAGAAACGAAAGCCACATTGGTGGCGGGTGCTCTGCACACGACTCGCTCCCTACCACACGCCCCCCGCTCCGCACACGACCGATCCGGGGACTGGAGCAGGAGGGCTGCACCGGGACTCCGGGACAGGCCCAGTTGAAAACGGCGGGGCGAGGGGGTGGGGTGGAGACGCCCGCGACGCCAAGGCTGGGGTCCCGGAAAGCGCGGGGAGGAGGGTGGAAGGCAAAGGCAACTTCGGGGAAACTGGGAAAGGCGGCCGGGACCTCGGGGACACTGCGTACCACCCGGCGCACAGCCCCTCCCGCGCAAACCCGAGCCAAAGGGGCGGTCGAGCGGCGCACTCGGAGCGGAGCTCAGGGGATGGTGCGCCCGGCACTTATGCTGGGCTCAGCCCAGCCCAGCCCAGGACCGCTGGCGGTGCGAACCCAGCCGGGCGGGTGCAAGCGCGGGGCAGGGCCTCTGCGCCCGGCCCCCTCCCCTGAGTAGAAAAGCAGCCGCAGGCTGTGGCGCTCCA
In mutant vector, “AC” highlighted were mutant to “TG”.
Supplementary
Acknowledgments
The authors would like to thank Yong-Fei Tan for the experimental design and instructions.
Funding: This study was supported in part by a grant from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.02.61). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin 2014;64:252-71. [Crossref] [PubMed]
- Watanabe SI, Nakagawa K, Suzuki K, et al. Neoadjuvant and adjuvant therapy for Stage III non-small cell lung cancer. Jpn J Clin Oncol 2017;47:1112-18. [Crossref] [PubMed]
- Tabchi S, Kassouf E, Rassy EE, et al. Management of stage III non-small cell lung cancer. Semin Oncol 2017;44:163-77. [Crossref] [PubMed]
- Katoh M. Combination immuno-oncology therapy with pembrolizumab, an anti-PD-1 monoclonal antibody targeting immune evasion, and standard chemotherapy for patients with the squamous and non-squamous subtypes of non-small cell lung cancer. J Thorac Dis 2018;10:5178-83. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-28. [Crossref] [PubMed]
- Coyle P, Philcox JC, Carey LC, et al. Metallothionein: the multipurpose protein. Cell Mol Life Sci 2002;59:627-47. [Crossref] [PubMed]
- Kumari MV, Hiramatsu M, Ebadi M. Free radical scavenging actions of metallothionein isoforms I and II. Free Radic Res 1998;29:93-101. [Crossref] [PubMed]
- Krężel A, Maret W. The Functions of Metamorphic Metallothioneins in Zinc and Copper Metabolism. Int J Mol Sci 2017; [Crossref] [PubMed]
- Calvo J, Jung H, Meloni G. Copper metallothioneins. Iubmb Life 2017;69:236-45. [Crossref] [PubMed]
- Floriańczyk B, Osuchowski J, Kaczmarczyk R, et al. Influence of metallothioneins on zinc and copper distribution in brain tumours. Folia Neuropathol 2003;41:11-14. [PubMed]
- Ohba KI. Transport and Toxicity of Cadmium. Nihon Eiseigaku Zasshi 2018;73:269-74. [Crossref] [PubMed]
- Zhang H, Lv S, Xu H, et al. H(2)O(2) Is Involved in the Metallothionein-Mediated Rice Tolerance to Copper and Cadmium Toxicity. Int J Mol Sci 2017; [Crossref] [PubMed]
- Werynska B, Pula B, Kobierzycki C, et al. Metallothioneins in the lung cancer. Folia Histochem Cytobiol 2015;53:1-10. [Crossref] [PubMed]
- Bizoń A, Jedryczko K, Milnerowicz H. The role of metallothionein in oncogenesis and cancer treatment. Postepy Hig Med Dosw (Online) 2017;71:98-109. [Crossref] [PubMed]
- Ji XF, Fan YC, Gao S, et al. MT1M and MT1G promoter methylation as biomarkers for hepatocellular carcinoma. World J Gastroenterol 2014;20:4723-29. [Crossref] [PubMed]
- Ding J, Lu SC. Low metallothionein 1M expression association with poor hepatocellular carcinoma prognosis after curative resection. Genet Mol Res 2016; [Crossref] [PubMed]
- Fu CL, Pan B, Pan JH, et al. Metallothionein 1M suppresses tumorigenesis in hepatocellular carcinoma. Oncotarget 2017;8:33037-46. [PubMed]
- Zhang S, Huang Z, Zhou S, et al. The effect and mechanism of metallothionein MT1M on hepatocellular carcinoma cell. Eur Rev Med Pharmacol Sci 2018;22:695-701. [PubMed]
- Chen Y, Quan R, Bhandari A, et al. Low metallothionein 1M (MT1M) is associated with thyroid cancer cell lines progression. Am J Transl Res 2019;11:1760-70. [PubMed]
- Hata A, Nakajima T, Matsusaka K, et al. A low DNA methylation epigenotype in lung squamous cell carcinoma and its association with idiopathic pulmonary fibrosis and poorer prognosis. Int J Cancer 2020;146:388-99. [Crossref] [PubMed]
- Changjun L, Feizhou H, Dezhen P, et al. MiR-545-3p/MT1M axis regulates cell proliferation, invasion and migration in hepatocellular carcinoma. Biomed Pharmacother 2018;108:347-54. [Crossref] [PubMed]
- Cao D, Ng TK, Yip Y, et al. p53 inhibition by MDM2 in human pterygium. Exp Eye Res 2018;175:142-47. [Crossref] [PubMed]
- Krizkova S, Fabrik I, Adam V, et al. Metallothionein--a promising tool for cancer diagnostics. Bratisl Lek Listy 2009;110:93-97. [PubMed]
- Cherian MG, Jayasurya A, Bay BH. Metallothioneins in human tumors and potential roles in carcinogenesis. Mutat Res 2003;533:201-9. [Crossref] [PubMed]
- Si M, Lang J. The roles of metallothioneins in carcinogenesis. J Hematol Oncol 2018;11:107. [Crossref] [PubMed]
- Basu A, Krishnamurthy S. Cellular responses to Cisplatin-induced DNA damage. J Nucleic Acids 2010; [Crossref] [PubMed]
- Jadhav RR, Ye Z, Huang RL, et al. Genome-wide DNA methylation analysis reveals estrogen-mediated epigenetic repression of metallothionein-1 gene cluster in breast cancer. Clin Epigenetics 2015;7:13. [Crossref] [PubMed]
- Denissenko MF, Pao A, Tang M, et al. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science 1996;274:430-32. [Crossref] [PubMed]
- Niazi S, Purohit M, Niazi JH. Role of p53 circuitry in tumorigenesis: A brief review. Eur J Med Chem 2018;158:7-24. [Crossref] [PubMed]
- Dong X, Ding W, Ye J, et al. MiR-24-3p enhances cell growth in hepatocellular carcinoma by targeting metallothionein 1M. Cell Biochem Funct 2016;34:491-6. [Crossref] [PubMed]
- Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004;303:844-8. [Crossref] [PubMed]
- Purvis JE, Karhohs KW, Mock C, et al. p53 dynamics control cell fate. Science 2012;336:1440-4. [Crossref] [PubMed]
- Felix-Portillo M, Martinez-Quintana JA, Arenas-Padilla M, et al. Hypoxia drives apoptosis independently of p53 and metallothionein transcript levels in hemocytes of the whiteleg shrimp Litopenaeus vannamei. Chemosphere 2016;161:454-62. [Crossref] [PubMed]
- Ostrakhovitch EA, Song YP, Cherian MG. Basal and copper-induced expression of metallothionein isoform 1,2 and 3 genes in epithelial cancer cells: The role of tumor suppressor p53. J Trace Elem Med Biol 2016;35:18-29. [Crossref] [PubMed]