
MAFA-AS1, a long non-coding RNA, predicts for poor survival of hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is one of leading cancers worldwide (50% in China alone). It remains the second cause of death resulted from cancer (1). In China, HCC was the third most common cancer and the second most lethal tumor (2). Although there have been advancements in diagnosis and treatment recently, only a small group of patients receiving surgeries are completely relieved. Given the poor prognosis of patients with HCC, more precise and detailed work is indispensable. As we know, carcinogenesis is a multi-step and multi-factor driven process (3). In addition to some well-known factors [e.g. hepatitis B virus (HBV) infection or alcoholism], micro-RNAs (miRNAs), long non-coding RNAs (lncRNAs) and genetic alterations also contribute to HCC tumorigenesis and progression (4,5).
lncRNAs are a group of non-coding RNAs that are considered to be longer than 200 nucleotides (6). They make up most of our genome and was recognized as useless noises, since they are not able to produce proteins (7). However, expression profiles of lncRNA bear tissue specificities, which means they are not functionally redundant (6,8). Actually, lncRNAs could act as transcription co-factors to regulate expression of adjacent protein-coding genes, interact with RNA binding proteins (RBPs) and regulate genes in cis, as well as affect epigenetics via chromatin modification (8,9). Other evidences implied that aberrant expression of lncRNAs was involved in pathological conditions like lung cancer, HCC and other cancers (10,11). Recent studies indicate that some exosomal lncRNAs are promising biomarkers for many cancer (12). Micro-RNAs belong to small non-coding RNA, which is about 21–22 nucleotides in length (13). Deregulation of miRNAs could become either engine or brake of oncogenesis (14). Previous literatures have shown that both lncRNAs and miRNAs could be key regulators in cancer stem cell differentiation and self-renew ability (15).
A variety of lncRNAs have been proved to be aberrantly expressed in tumor, for example, Yan et al. found the imbalanced up-regulation of H19 in gastric cancer tissues (16). Recently, some lncRNAs have been recognized as novel prognostic biomarkers of tumor, for example, lncRNA GAS5 expression is higher in non-small cell lung cancer (NSCLC) tumor tissues than in normal tissues, and high expression of GAS5 was correlated with advanced clinical stage and poorer outcome (17). Additionally, many studies showed that abnormal expression of lncRNAs such as H19, HOTAIR and MALAT1 might contribute to tumor development (16,18,19). Many lncRNAs promote or inhibit carcinogenesis via interacting with mRNAs (20). MiRNAs can conditionally bind lncRNA on specific site and reduce the decay of target genes (21). Since complicated interaction exist among lncRNA, miRNAs and associated proteins of HCC, a more detailed visualization of the network is necessary. In this study, some candidate oncogenic or tumor-suppressive lncRNAs in HCC were screened by integrative bioinformatics analyses. MAFA-AS1 was identified as a candidate lncRNA because of its prognostic value for overall and disease-free survival (DFS) of patients with HCC. It also has potential to interact with SRSF1 and/or SRSF9 and affect DNA replication and cell cycle. Additionally, MAFA-AS1 could interact with miR210, an important oncogenic miRNA in many cancers (22,23). The molecular interactions among lncRNAs, miRNAs and associated proteins were explored.
Methods
Statement of ethics approval
The Statement of Ethics Approval is not required because this is a bioinformatics study.
Screening of aberrantly expressed and genomically altered lncRNAs in HCC samples
The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/) is a public cancer database of genomic and clinical information containing RNA-seq, single-nucleotide variations (SNVs) and copy number variations (CNVs), in 33 kinds of malignant tumors. Firstly, RNA-Seq-HTseq- count raw data of LIHC including 371 HCC tissues and 50 liver tissues was downloaded from TCGA (24). The raw data were inputted into R to analyze and visualize the aberrantly expressed lncRNAs. Fold change (FC) >2 (log2FC>1) and P<0.05 between tumor and non-tumor tissues were considered as significant. As further narrow down the threshold to log2FC >1.5, the eligible lncRNAs were screened by edgeR package and limma package, respectively. Two lists of altered lncRNAs were integrated, and finally 1384 lncRNAs were achieved. These lncRNAs were then imported into DAVID (www.DAVID.com) and HGNC (www.genenames.org) to get gene symbol of each lncRNA (25,26).
To acquire those officially acknowledged lncRNAs in transcriptome and genome simultaneously, the most differentially expressed officially approved lncRNAs were imported into the OncoPrint module of cBioportal (http://www.cBioportal.org) for genomic analysis (27,28). The lncRNAs changed in more than 5% of the cases were taken as remarkable ones. The overlapping of genomic altered lncRNAs and the abnormally expressed lncRNAs (log2FC>2, P<0.05) are implemented by VENNY (http://bioinfogp.cnb.csic.es/tools/venny/index.html). Hierarchical cluster analyses are performed by R in ggplot2 package (29).
Clinical properties and survival analysis in HCC
To further investigate the association between lncRNAs and the Clinicopathologic characters (survival, stage and grade, etc.), GEPIA (http://gepia.cancer-pku.cn/) was applied to analyze the survival and stages of patients with lncRNAs based on TCGA RNA-Seq data (30). CBioportal, another online tool, was used for analyzing survival data of HCC cases with or without CNVs (27,28). Visualization of overall survival (OS), DFS and stages were analyzed by GEPIA and cBioportal. Correlations between tumor grades and lncRNAs were evaluated and visualized by TANRIC (http://ibl.mdanderson.org/tanric/_design/basic/index.html) (31). Kaplan-Meier method was used to generate survival curves. The association of lncRNAs with TNM stage or grade was evaluated by ANOVA or student t-test. P<0.05 was set as statistically significant.
Integration of co-expressed genes, RNA-protein interaction network, gene ontology/pathway enrichment analysis and associated miRNAs
In order to probe the underlying molecular mechanism of MAFA-AS1 in HCC, we took advantage of circlncRNA database (http://120.126.1.61/circlnc/circlncRNAnet/lncRNA_TCGA/index.php) to calculate the co-expressed genes of MAFA-AS1 along with their chromosomal locations. Co-expressed genes with Pearson P value below 0.05 on the basis of TCGA LIHC samples were regarded as significant. Besides, we conducted GO/KEGG analysis and RBPs were predicted by circlncRNAnet (32). In addition to looking for related lncRNAs in TANRIC, we utilized LinkedOmics website (http://www.linkedomics.org/login.php) and GEPIA to analyze the survival information of patients according to profiles of the RBPs and miRNAs.
Results
Aberrantly expressed lncRNAs in TCGA LIHC samples
Raw data of TCGA LIHC was inputted into R and differentially expressed lncRNAs were analyzed. Following the criteria (log2FC >1, P<0.05), there are 1,932 lncRNAs considered to be dysregulated between tumor and nontumor tissues, with 1722 upregulated and 210 downregulated lncRNAs (Figure 1A). With log2FC >1.5 (P<0.05) threshold, 1,384 lncRNAs were enrolled into our study. These lncRNAs were integrated into DAVID (www.DAVID.com) and HGNC (www.genenames.org) to blast with official gene symbols, only 454 symbols were acknowledged by HGNC. Indeed, 230 acknowledged lncRNAs in total conformed to a more stringent standard (log2FC >2).
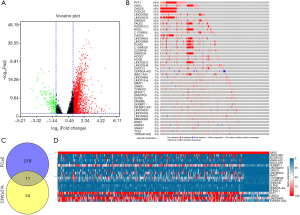
LncRNAs with highest frequency in CNVs
Genomic changes tend to play a key role in many tumors including HCC. We looked for meaningful mutations in genome, but no valuable lncRNA mutation was observed. However, the CNVs, including amplification and deep deletion, were found in those lncRNAs. CNV information of 453 lncRNAs was obtained, except AC005150.1, whose data was not available in cBioportal. There are 45 of 453 lncRNAs were altered in more than 5% of the HCC patients according to cBioportal. Among them, PVT1 had the highest frequency (24%) and most lncRNAs amplified rather than deleted (Figure 1B).
LncRNAs are systematically dysregulated in genomics and transcriptomics in HCC
It is believed that lncRNAs with concurrent changes in transcriptome and genome might be critical in HCC. We extracted 230 differentially expressed lncRNAs mentioned above and 45 lncRNAs with obvious CNVs. By integrating bioinformatics analysis in VENNY, a total of 11 lncRNAs including CDKN2A-AS, BPESC1, ELFN2, CASC9, C17ORF82, RMST, TSPEAR-AS2, PVT1, LINC00200, C2ORF48, GUSBP11, were identified from the datasets (Figure 1C). Ten most differentially expressed lncRNAs and 10 most frequently CNV-altered lncRNAs were also included for gene expression analysis (Figure 1D).
LncRNAs correlate to poor survival rates of patients with HCC
Twenty lncRNAs were retrieved in GEPIA and CBioportal to validate the correlation between lncRNAs and the clinical outcomes based on TCGA database. Among them, the high expression of LINC00200, MAFA-AS1, CASC8, CASC9, LINC01667, CDKN2A-AS1 indicates poor five-year OS rate in HCC (Figure 2A). Kaplan-Meier analysis revealed that high expression of MAFA-AS1, CASC9, LINC01667, CDKN2A-AS1 were significantly correlated with poorer DFS rates of HCC patients (Figure 2B). With the help from CBioportal, we also analyzed the survival data of those lncRNAs to determine whether HCC patients with genomic alterations in those lncRNAs had poor survival outcome. However, no association between the CNVs of the lncRNAs and survival has been found.
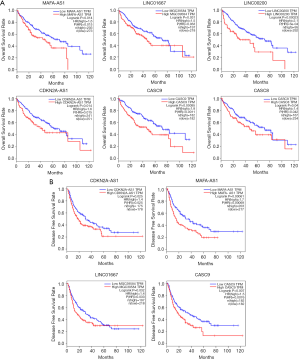
MAFA-AS1 is a candidate biomarker for poor prognosis of HCC patients
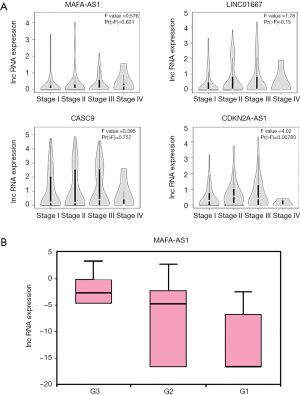
The clinical-pathological properties also reflect for prognosis in HCC (33). We used GEPIA and TANRIC to determine the correlations between the lncRNAs and clinical characters of HCC. The profile of MAFA-AS1 was able to distinguish early-stage HCC patients from advanced stage patients (Figure 3A). The other three lncRNAs (LINC01667, CASC9, CDKN2A-AS1) showed positive correlation with stages except stage IV (Figure 3A). Besides, high expression of MAFA-AS1was observed in higher tumor grade, which means loss of differentiation, a sign of extremely malignancy (Figure 3B). Taken together, these results indicate that MAFA-AS1 might be a novel prognostic marker for HCC.
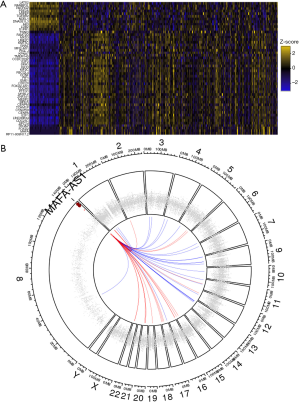
MAFA-AS1 interacts with SRSF1/SRSF9 and miR210
Because MAFA-AS1 high expression was significantly associated with worse outcome in HCC, the role of MAFA-AS1 in HCC progression was further explored. Recent studies have revealed that one of the major functions of lncRNA was to mediate the expression of nearby genes (8). We were wondering whether some oncogenes or tumor suppressors were regulated by MAFA-AS1. We evaluated certain genes correlating with MAFA-AS1 in HCC by TCGA co-occurrence analyses (circlncRNAnet). Fifty most correlated co-expressing genes based on TCGA LIHC samples were presented by heatmap (Figure 4A). The chromosomal locations of these genes are presented by Circos plot. As shown in Figure 4B, the co-expressing genes are randomly located on almost all chromosomes except chromosome X and Y. The locations of those genes are far from the MAFA-AS1 locus, suggesting that MAFA-AS1 might not affect those genes via space interactions.
LncRNAs were reported to bind to RBPs as partner and adjust stability of those proteins only on the condition that lncRNAs have a considerable firm structural conformation (34). We next characterized the secondary structure of MAFA-AS1 in RNAfold web server. As Figure 5A shown, MAFA-AS1 has a stable secondary structure. Through analysis of related RBPs in circlncRNAnet, SRSF1 and SRSF9 were predicted to interact with MAFA-AS1 (Figure 5B). Survival analyses suggested that high expression of SRSF1or SRSF9 indicated poor OS and DFS (Figure 5C,D). SRSF1/SRSF9 was reported to play a key role in HCC progression by modulating DNA damage repair mechanism and cell cycle (35). Additionally, MAFA-AS1 mainly affects DNA replication and cell cycle by GO and KEGG analyses (Figure 5E,F).
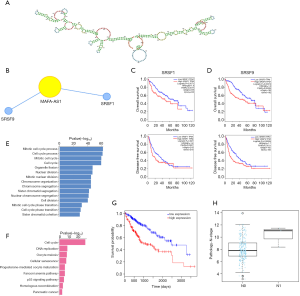
Besides, it was also predicted that MAFA-AS1 correlated with hsa-miR-210 via TANRIC (R=0.401, P<0.001). MiR210 is a hypoxia specific miRNA participating in many cancers (23). MiR210 high expression predicts for poor survival of patients with HCC (Figure 5G). It is significantly upregulated in HCC patients with lymphoid node metastasis (Figure 5H).
Discussion
Although lots of studies were conducted to investigate the mechanism of HCC tumorigenesis and progress, it remains perplexing and contradictory. One reason is that most results were generated from a single cohort and/or small sample study. Integrating the genomic and transcriptomic information from multiple databases and conducting bioinformatics analyses is a practical way to find potential biomarkers and therapeutic targets in HCC tumorigenesis and progression. Many lncRNAs were reported to be correlated with clinical outcomes of HCC patients (36). However, considering the complex factors and mechanisms accounting for HCC tumorigenesis, more detailed and accurate analyses were necessary. Here, we identified a cluster of upregulated lncRNAs (MAFA-AS1, CASC9, LINC01667, CDKN2A-AS1) as indicators for poor prognosis of HCC patients by data-mining LIHC (a TCGA HCC database). One of these lncRNAs, MAFA-AS1, can distinguish early-stage HCC patients from advanced stage patients. The high expression of MAFA-AS1 predicts for poor OS and disease-free survival of HCC patients. Instead of co-acting with the neighboring genes, MAFA-AS1 interacts with RBPs SRSF1 or SRSF9 and it might affect DNA replication and cell cycle. Both SRSF1 and SRSF9 are members of the SR (splicing regulators) protein family (37). SRSF1 is involved in some key parts of mRNA metabolism (for example, mRNA splicing, mRNA stability and mRNA translation) and other processes (for instance, nucleolar stress response, protein sumoylation and miRNA processing) (37). Moreover, SRSF1 and SRSF9 were reported previously to activate Wnt signaling pathways by increasing biosynthesis of β-catenin (38). MAFA-AS1 might exert oncogenic effect in HCC by interacting with SRSF1/SRSF9 via being scaffold. MAFA-AS1 was also positively correlated with miR210, an onco-miRNA that predicts for poor OS in patients with HCC.
The combination of multiple bioportals with TCGA database provides an efficient way to seek out lncRNAs that can predict clinical outcome. It opens a window to elucidate the possible molecular mechanisms of oncogenic role of lncRNAs in HCC. Our study demonstrates for the first time that MAFA-AS1 is a candidate lncRNA for HCC carcinogenesis via binding splicing factors SRSF1/SRSF9 and co-acting with miR210. Unlike other studies that lncRNA affects tumor progression mainly via the mediation of connected genes (39), our research displays a more comprehensive network of lncRNA-RBPs and miRNAs modulation.
Acknowledgments
We thank the website (https://www.shengxin.ren/) and its engineer Rangfei Zhu for the guidance in bioinformatics. We also thank Dr. Lei Li for assistance on the manuscript.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.03.11). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The Statement of Ethics Approval is not required because this is a bioinformatics study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Zuo TT, Zheng RS, Zhang SW, et al. Incidence and mortality of liver cancer in China in 2011. Chin J Cancer 2015;34:508-13. [Crossref] [PubMed]
- Ventura A, Kirsch DG, McLaughlin ME, et al. Restoration of p53 function leads to tumour regression in vivo. Nature 2007;445:661-5. [Crossref] [PubMed]
- Su CH, Lin Y, Cai L. Genetic factors, viral infection, other factors and liver cancer: an update on current progress. Asian Pac J Cancer Prev 2013;14:4953-60. [Crossref] [PubMed]
- Xiong H, Ni Z, He J, et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene 2017;36:3528-40. [Crossref] [PubMed]
- Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov 2017;16:167-79. [Crossref] [PubMed]
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. [Crossref] [PubMed]
- Wei JW, Huang K, Yang C, et al. Non-coding RNAs as regulators in epigenetics Oncol Rep 2017;37:3-9. (Review). [Crossref] [PubMed]
- Wang X, Arai S, Song X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 2008;454:126-30. [Crossref] [PubMed]
- Terashima M, Tange S, Ishimura A, et al. MEG3 Long Noncoding RNA Contributes to the Epigenetic Regulation of Epithelial-Mesenchymal Transition in Lung Cancer Cell Lines. J Biol Chem 2017;292:82-99. [Crossref] [PubMed]
- Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol 2018;15:137-51. [Crossref] [PubMed]
- Dragomir M, Chen B, Calin GA. Exosomal lncRNAs as new players in cell-to-cell communication. Transl Cancer Res 2018;7:S243-52. [Crossref] [PubMed]
- Varrone F, Caputo E. The miRNAs Role in Melanoma and in Its Resistance to Therapy. Int J Mol Sci 2020; [Crossref] [PubMed]
- Prabhu KS, Raza A, Karedath T, et al. Non-Coding RNAs as Regulators and Markers for Targeting of Breast Cancer and Cancer Stem Cells. Cancers (Basel) 2020; [Crossref] [PubMed]
- Huang T, Alvarez A, Hu B, et al. Noncoding RNAs in cancer and cancer stem cells. Chin J Cancer 2013;32:582-93. [Crossref] [PubMed]
- Yan J, Zhang Y, She Q, et al. Long Noncoding RNA H19/miR-675 Axis Promotes Gastric Cancer via FADD/Caspase 8/Caspase 3 Signaling Pathway. Cell Physiol Biochem 2017;42:2364-76. [Crossref] [PubMed]
- Tan Q, Zuo J, Qiu S, et al. Identification of circulating long non-coding RNA GAS5 as a potential biomarker for non-small cell lung cancer diagnosisnon-small cell lung cancer, long non-coding RNA, plasma, GAS5, biomarker. Int J Oncol 2017;50:1729-38. [Crossref] [PubMed]
- Tao D, Zhang Z, Liu X, et al. LncRNA HOTAIR promotes the invasion and metastasis of oral squamous cell carcinoma through metastasis-associated gene 2. Mol Carcinog 2020;59:353-64. [Crossref] [PubMed]
- Kim J, Piao HL, Kim BJ, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet 2018;50:1705-15. [Crossref] [PubMed]
- Mazza T, Mazzoccoli G, Fusilli C, et al. Multifaceted enrichment analysis of RNA-RNA crosstalk reveals cooperating micro-societies in human colorectal cancer. Nucleic Acids Res 2016;44:4025-36. [Crossref] [PubMed]
- Zheng J, Huang X, Tan W, et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat Genet 2016;48:747-57. [Crossref] [PubMed]
- Liu SS, Chan KKL, Chu DKH, et al. Oncogenic microRNA signature for early diagnosis of cervical intraepithelial neoplasia and cancer. Molecular oncology 2018;12:2009-22. [Crossref] [PubMed]
- Yu P, Fan S, Huang L, et al. MIR210 as a potential molecular target to block invasion and metastasis of gastric cancer. Med Hypotheses 2015;84:209-12. [Crossref] [PubMed]
- Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68-77. [Crossref] [PubMed]
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc 2009;4:44-57. [Crossref]
- Huang da W. Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1-13. [Crossref] [PubMed]
- Lee H, Palm J, Grimes SM, et al. The Cancer Genome Atlas Clinical Explorer: a web and mobile interface for identifying clinical-genomic driver associations. Genome Med 2015;7:112. [Crossref] [PubMed]
- Gong L, Zhang D, Dong Y, et al. Integrated Bioinformatics Analysis for Identificating the Therapeutic Targets of Aspirin in Small Cell Lung Cancer. J Biomed Inform 2018;88:20-8. [Crossref] [PubMed]
- Ito K, Murphy D. Application of ggplot2 to Pharmacometric Graphics. CPT Pharmacometrics Syst Pharmacol 2013;2:e79. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucl Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Li J, Han L, Roebuck P, et al. TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Cancer Res 2015;75:3728-37. [Crossref] [PubMed]
- Wu SM, Liu H, Huang PJ, et al. circlncRNAnet: An integrated web-based resource for mapping functional networks of long or circular forms of non-coding RNAs. Giga Science 2018;7:1-10.
- Cai MY, Wang FW, Li CP, et al. Prognostic factors affecting postoperative survival of patients with solitary small hepatocellular carcinoma. Chin J Cancer 2016;35:80. [Crossref] [PubMed]
- Ferrè F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform 2016;17:106-16. [Crossref] [PubMed]
- Chen L, Luo C, Shen L, et al. SRSF1 Prevents DNA Damage and Promotes Tumorigenesis through Regulation of DBF4B Pre-mRNA Splicing. Cell Rep 2017;21:3406-13. [Crossref] [PubMed]
- Zhang M, Wang W, Li T, et al. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother 2016;80:73-9. [Crossref] [PubMed]
- Das S, Krainer AR. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol Cancer Res 2014;12:1195-204. [Crossref] [PubMed]
- Fu Y, Huang B, Shi Z, et al. SRSF1 and SRSF9 RNA binding proteins promote Wnt signalling-mediated tumorigenesis by enhancing beta-catenin biosynthesis. EMBO Mol Med 2013;5:737-50. [Crossref] [PubMed]
- Gong H, Wen H, Zhu X, et al. High expression of long non-coding RNA ZEB1-AS1 promotes colorectal cancer cell proliferation partially by suppressing p15 expression. Tumour Biol 2017;39:1010428317705336. [Crossref] [PubMed]


